
Hellnardos at 13th GSCN conference @gscnoffice.bsky.social. We are excited about the next 3 days of stem cell science! 🧫
15.10.2025 09:02 — 👍 3 🔁 1 💬 0 📌 0@felixpfoertner.bsky.social
PhD student @enardhellmannwg.bsky.social

Hellnardos at 13th GSCN conference @gscnoffice.bsky.social. We are excited about the next 3 days of stem cell science! 🧫
15.10.2025 09:02 — 👍 3 🔁 1 💬 0 📌 0
🎉Excited to share our new preprint: “Improving RNA-seq protocols”
We introduce a systematic approach to maximize usable reads in RNA-seq.
With this, we created prime-seq2, improving one of the most cost-efficient bulk RNAseq protocols.
👉Check it out: doi.org/10.1101/2025...
Huge thanks to Eva (shared first author), and to Wolfgang & Daniel, who supervised the project 🙏
🧵9/9
What became very clear throughout the project is how complex and unpredictable a, on the face of it, simple protocol can be. I think we mostly underestimate the complexity of molecular protocols.
Also: there’s room for discussion and research into what we consider “usable” reads (discussion).🧵8/9
We think our “funnel strategy” of maximizing usable reads could also be applied more broadly — as a QC metric and as a systematic way to optimize other bulk and single-cell RNA-seq protocols.🧵7/9
26.08.2025 14:38 — 👍 0 🔁 0 💬 1 📌 0
We combined these changes into prime-seq2. Across samples and libraries, it delivers +60% usable reads compared to prime-seq.
If you’d like to try it: we published a detailed, step-by-step protocol 🧪
👉 dx.doi.org/10.17504/pro...
🧵6/9

For prime-seq, the main loss points were:
• total sequencing reads
• index assignment
• intergenic reads
To improve this, we tested many alterations and ended up with:
✅ optimized DNA digest
✅ introduced PTO bonds into PCR primers
✅ redesigned ligation adapters & library PCR primers🧵5/9

This process can be nicely visualized as a funnel: at each step, reads are lost. The funnel makes it clear which stages are most responsible for read loss — and therefore where improvements could make the biggest impact.🧵4/9
26.08.2025 14:38 — 👍 0 🔁 0 💬 1 📌 0We think what matters is the number of reads that can be used for analysis, "usable reads", while controlling for traditional QC metrics.
The usable reads number depends on:
- valid reads from the flow cell
- correct index
- filtering
- correct barcode
- exonic/intronic assignment
- unique UMI🧵3/9
Prime-seq has been used extensively in our lab. Over time, several imperfections became clear. During my Master’s thesis, I started on testing many potential improvements. To decide which alterations to keep, we needed a clear QC metric.🧵2/9
26.08.2025 14:38 — 👍 0 🔁 0 💬 1 📌 0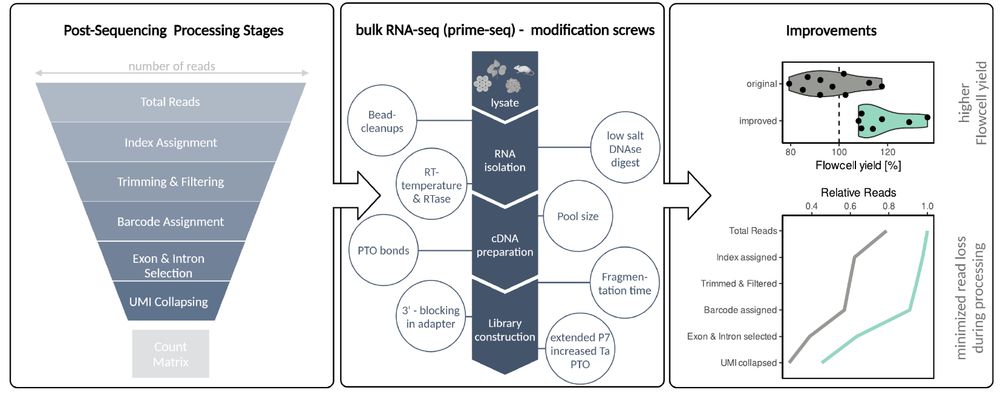
I am thrilled to finally share our new preprint on prime-seq2 🚀
We improved one of the most cost-efficient bulk RNAseq protocols out there to end up with +60% usable reads.
Check it out: doi.org/10.1101/2025...
a🧵1/9

This is the beginning of our office introduction series (1/5):
Meet the office of Leo, Antonia with Wilma and Felix
Leo 🧠 maps FOXP2 brains
Antonia 🧫 reprograms cross-species cells
Felix ⚙️ drives Part 2 of every project
Wilma 🐶 heads cable-integrity testing by chewing
#LabLife #BlueskyScience
The Enard/Hellmann group is now on bluesky 🦋
We are looking forward to reconnecting with everyone here and to welcoming new followers.
Let’s keep up great science interactions online!