Corush Lab
I'm recruiting a Ph.D. student for Fall 2026.
Interested in comparative studies and trait evolution in fishes?
Send me an email with a CV and research interests. Please take a look at my website (jcorush.github.io ) for more information about my research.
#hybridization #minnows #mudskippers
04.12.2025 00:38 — 👍 32 🔁 57 💬 1 📌 1
Fish Division | Museum of Biological Diversity
NEWS from Ohio State Fish Division: You can now search our 6,000-tissue collection on our website! 🐟
mbd.osu.edu/collections/...
12.12.2025 16:01 — 👍 32 🔁 14 💬 2 📌 0

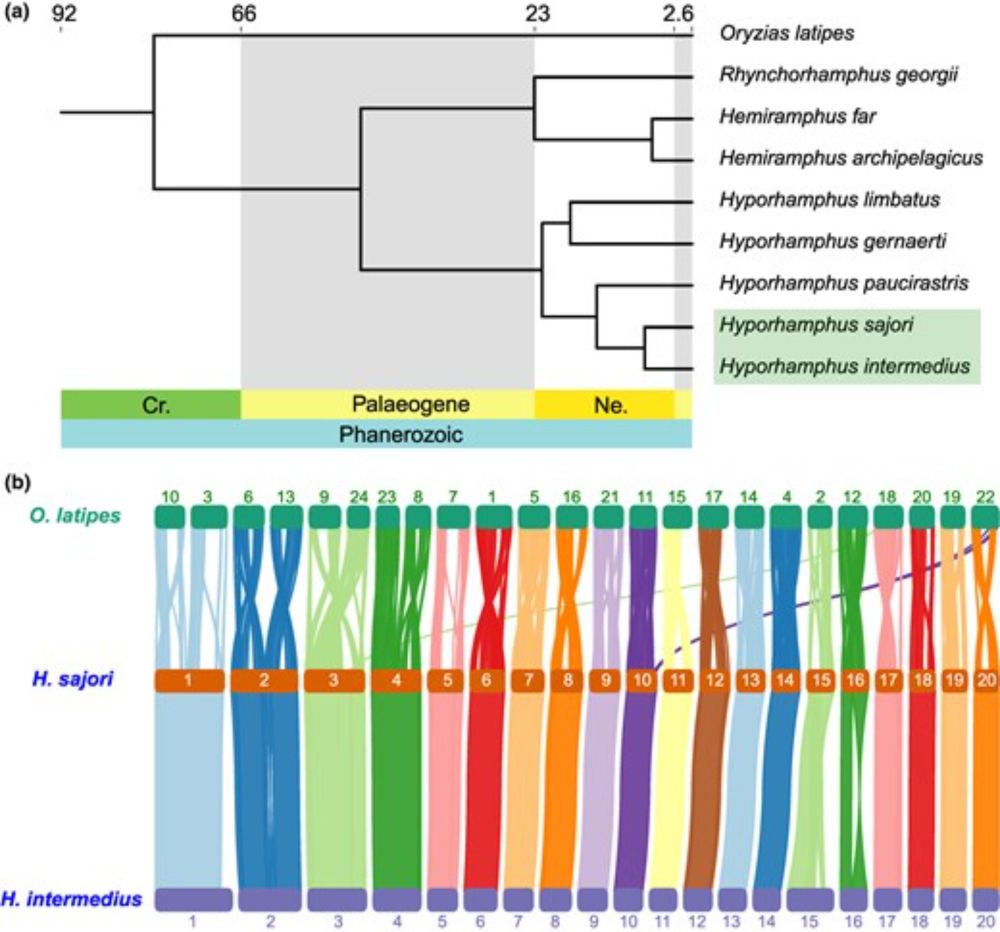
Promoting the use of phylogenetic multinomial generalised mixed-effects model to understand the evolution of discrete traits
Abstract. Phylogenetic comparative methods (PCMs) are fundamental tools for understanding trait evolution across species. While linear models are widely us
NEW METHODS ARTICLE: Phylogenetic GLMMs open doors to study evolution of discrete traits. We show how binary models extend to ordinal & nominal traits, using bird data, and provide tutorials to make these methods accessible to evolutionary biologists:
doi.org/10.1093/jeb/...
Mizuno et al.
11.12.2025 14:32 — 👍 25 🔁 10 💬 0 📌 0
You can do it using the Network Analysis toolbox in QGIS. There is a "shortest path" option. I recall there was a way to make a distance matrix between all of your point based on a specific layer, your rivers.
09.12.2025 17:36 — 👍 2 🔁 0 💬 0 📌 0
My lab focuses on:
-Nest association/breeding behavior evolution in N. American Minnows.
-Mudskipper evolution and biogeography.
But I am open to other related projects addressing #hybridization #PhylogeneticComparativeMethods #LandscapeGenetics #biogeography
email: jcorush @ Illinoistech. edu
08.12.2025 20:19 — 👍 0 🔁 1 💬 0 📌 0
@jcorush.bsky.social is looking for a PhD student
05.12.2025 23:21 — 👍 2 🔁 2 💬 0 📌 0
Corush Lab
I'm recruiting a Ph.D. student for Fall 2026.
Interested in comparative studies and trait evolution in fishes?
Send me an email with a CV and research interests. Please take a look at my website (jcorush.github.io ) for more information about my research.
#hybridization #minnows #mudskippers
04.12.2025 00:38 — 👍 32 🔁 57 💬 1 📌 1

Restoring the Klamath River - Tomorrow's Catch
Witness the Klamath River’s transformation as dam removal restores salmon habitat, tribal lands, and cultural connections in this historic restoration.
The Klamath River is flowing free again.
RES and local Indian Tribes are restoring habitat, renewing cultural ties, and leading the largest dam removal in history.
🎥 Watch the film + explore the series: contentwithpurpose.co.uk/afs/tomorrow...
20.11.2025 15:00 — 👍 14 🔁 6 💬 0 📌 0

Two BLM employees tag fish in Alaska.
A new report reveals that fish and wildlife conservation generates substantial economic benefits. Federal, state, local, and nonprofit contributions combined contributed $115.8 billion in total economic activity and supported over 575,000 jobs nationwide. Read more: fisheries.org/2025/11/new-...
19.11.2025 20:24 — 👍 12 🔁 6 💬 0 📌 1

MBE | Temperature and Pressure Shaped the Evolution of Antifreeze Proteins in Polar and Deep Sea Zoarcoid Fishes
A graphic visualizing the finding by Bogan et al. that antifreeze protein (AFP) genes increased in copy number among shallow-water, polar species of Zoarcoidei fishes. Top left: Pholis gunnellus photographed by Chris Isaacs (CC-BY-NC). Top right: Cebidichthys violaceus photographed by Alex Heyman (CC0 1.0). Bottom left: Lycenchelys sp. photographed by Julien Savoie (CC BY 4.0). Bottom right: Lycodes sp. photographed by Julien Savoie (CC BY 4.0).
@snbogan.bsky.social @notothentoma.bsky.social @scotthotaling.bsky.social @paulbfrandsen.bsky.social et al. explore the evolution of type III antifreeze proteins in deep sea zoarcoid fishes.
🔗 doi.org/10.1093/molbev/msaf219
#evobio #molbio
21.10.2025 09:43 — 👍 18 🔁 8 💬 0 📌 1

Submit your ideas to the 100% Wisconsin Fish contest | Wisconsin Sea Grant
The contest is seeking creative ideas on how to use all parts of Great Lakes fish.
There's more to fish than the fillet, and we need your ideas on how to use it!
Submit your suggestions on how to use the whole fish to the 100% Wisconsin Fish contest for a chance to win a #GreatLakes prize pack.
www.seagrant.wisc.edu/news/submit-...
30.09.2025 13:34 — 👍 5 🔁 2 💬 0 📌 0
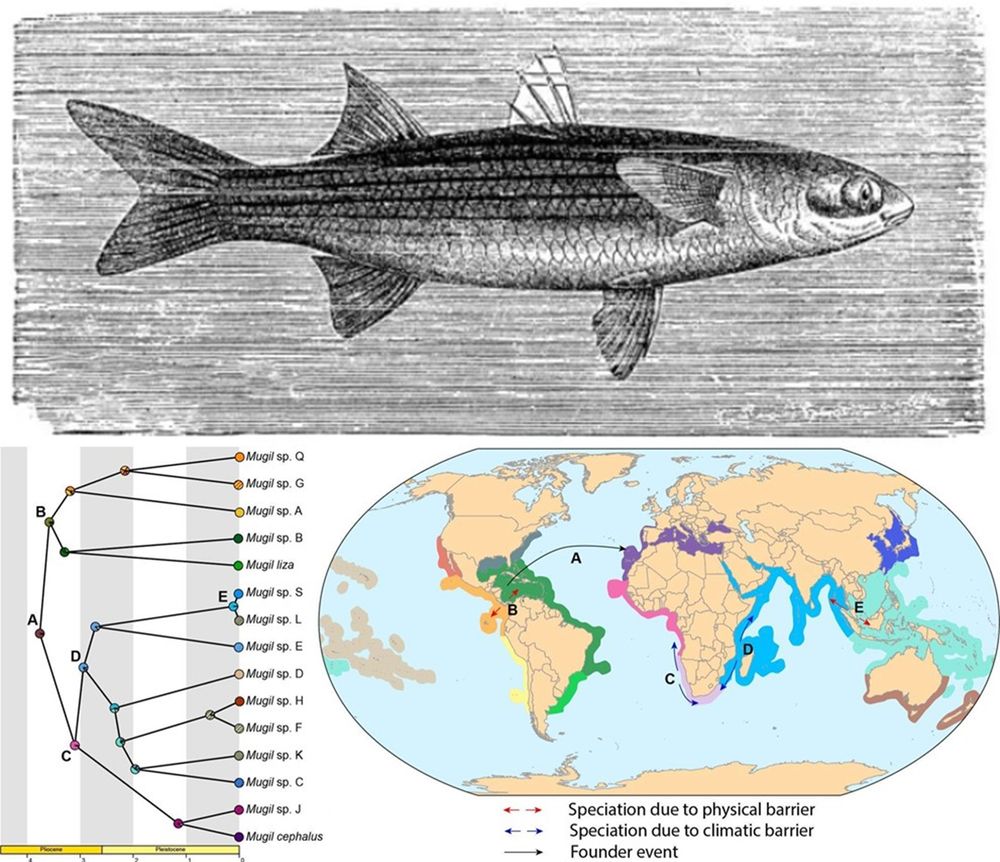
Non-anthropogenic cosmopolitan species ranges are scarce
Mugil cephalus, traditionally considered a (quasi)cosmopolitan diadromous fish species, is now shown to comprise some 15 different lineages deserving formal species description
www.sciencedirect.com/science/arti...
08.10.2025 08:45 — 👍 6 🔁 4 💬 0 📌 0

Unravelling the Effects of Ecology and Evolutionary History in the Phenotypic Convergence of Fishes
Abstract. Understanding the ecological drivers and limitations of adaptive convergence is a fundamental challenge. Here, we explore how adaptive convergenc
Super excited to share our new paper investigating convergence across planktivorous fishes and assessing the impact of ancestry and light environment led by Dr Jen Hodge @fishncurious.bsky.social and including most members of the lab past and present academic.oup.com/sysbio/advan...
14.05.2025 04:50 — 👍 55 🔁 21 💬 2 📌 1

Rapid Response Bridge Funding Program
In the face of recent abrupt shifts in federal funding for education research, including large-scale terminations of National Science Foundation (NSF) research grant awards, we have developed a rap...
**Rapid Response Bridge Funding Program** -- $25,000 grants earmarked for early-career researchers whose NSF-funded research on STEM and education has just been terminated.
www.spencer.org/grant_types/...
hub.jhu.edu/2025/04/28/j...
Thanks @lizneeley.bsky.social !
03.05.2025 15:22 — 👍 76 🔁 72 💬 0 📌 4

Fishes of the Chicago Region: A Field Guide
Buy Fishes of the Chicago Region: A Field Guide on Amazon.com ✓ FREE SHIPPING on qualified orders
New fish book alert! On this #FishFriday, "Fishes of the Chicago Region" was just released. Detailed info on species distributions & natural history observations for >150 species in SE Wisconsin, NE Illinois, northern Indiana, and SW Michigan. Check it out...
www.amazon.com/Fishes-Chica...
18.04.2025 16:52 — 👍 19 🔁 11 💬 0 📌 1
Congrats!!
14.04.2025 18:15 — 👍 1 🔁 0 💬 0 📌 0

Some microphagous fishes have a unique pocket-like throat structure, epibranchial organ (EBO), that helps gather tiny food particles. New study of 13 species reveals diverse EBO anatomy, weak phylogenetic ties & possible link to diet
Evans et al:
anatomypubs.onlinelibrary.wiley.com/doi/10.1002/...
03.04.2025 16:04 — 👍 22 🔁 15 💬 1 📌 3

(A) Likelihood of the top five best-fitting trees based on f2 statistics in 500 kb windows across the genome of the 6 bear populations studied (Ancient polar bear = APB, modern polar bears = PB, Alaskan brown bears = BB, Baranof and Chichagof brown bears = ABC-BC, European brown bear = EBB, American black bear = BLK). Tree A is the best fit and reflects the expected genetic relationships among the bear populations. Trees D and E are the fourth and fifth best-fitting trees reflecting hybridization from polar bears into brown bears (Tree D) and vice versa (Tree E). (B) Cases of hybridization among the best-fitting trees in (A) (trees D and E) and cases of incomplete lineage sorting (trees L and J) which fit worse than trees D and E.
Polar bears and their lower-latitude cousins—brown bears—have a complicated past! Genomes from a >115,000 year old polar bear and 65 modern bears reveal that although polar bears and brown bears began diverging >1 mya, they hybridized up until ~200 kya. #2025MMM doi.org/10.1073/pnas.2200016119
25.03.2025 00:40 — 👍 38 🔁 12 💬 3 📌 0

Fig. 1: The Chromosome number and Hidden State-dependent Speciation and Extinction (ChromoHiSSE) model. Panel (a) describes the event rates allowed in the model for both cladogenetic (upper) and anagenetic (lower) events. Panels (b) and (c) demonstrate those rates on a tree simulated under ChromoHiSSE. Branches that do not reach the present represent extinct, unsampled lineages. Panel (b) shows the anagenetic and cladogenetic changes in the hidden states, indicated by blue (i) vs orange (ii). Panel (c) shows the anagenetic and cladogenetic gains and losses of chromosomes, as well as speciation with no corresponding change in chromosome numbers. More red colors in (c) correspond to more chromosomes. The vertical blue and orange bars in (c) indicate clades in the blue (i) vs orange (ii) hidden states, displaying that chromosome number changes are less frequent in the blue hidden state than in the orange. The asterisk in (c) demarcates where a cladogenetic dysploidy event could appear as an anagenetic event because of unsampled lineages.
New study by @tribblelab.bsky.social @jimarcor.bsky.social @marcialescudero.bsky.social Michael May, Rosana Zenil-Ferguson and myself just out:
Introduces a novel HiSSE chromosome model and demonstrates the importance of chrom. evol. in #sedge diversity.
nph.onlinelibrary.wiley.com/doi/10.1111/...
26.12.2024 13:01 — 👍 75 🔁 24 💬 1 📌 1
No.
LG statistically tests specific landscape-based resistance/barrier models on gene flow at an individual/population level. PG is a clade based understanding of the species' range. LG can inform PG, but PG does not have to incorporate landscapes into the hypotheses that are being tested.
17.12.2024 05:21 — 👍 3 🔁 0 💬 0 📌 0
Let´s form a community of conservation geneticists here - I started with some people I know, but there are likely to be many more... I can happily add them to this list
go.bsky.app/VMqVSAi
02.11.2024 08:19 — 👍 85 🔁 54 💬 54 📌 8
For scientists interested in large lakes of the world. Contact us if you'd like to be added. Space still available.
15.11.2024 02:16 — 👍 88 🔁 31 💬 26 📌 6
Assistant Professor at Wageningen University | Evolutionary Biology, Genomics and Ornithology | Blogging at Avian Hybrids
Parent, husband, ichthyologist, professor, Head Saybrook College,
@Yale_EEB chair, FirstGen, Chicagoan, New Havenite. www.nearlab.org/
Evolutionary biologist trying to do a bit of good | Ongoing #fishgenomics & #lichens work | Evolution, philosophy, pedagogy, fungi, genomics | Justice & climate | she/her
fish evolutionary biologist, data nerd, outdoor enthusiast, herder of small humans. Associate Prof. Views my own.
The Society for Freshwater Science (SFS) is an international scientific society whose purpose is to promote understanding of freshwater ecosystems. #2026SFS https://freshwater-science.org/
Curator of Herpetology & Associate Professor of Vertebrate Zoology at Natural History Museum of Denmark • ERC StG: GEMINI • Co-host of SquaMates Podcast and AnatomyInsights on Youtube • He/Him
Posdoctoral researcher 🔥| Forest Ecologist | Fire Ecology | Plant Flammability | Ecological Restoration | Seed Germination | Invasive Species
Professor of Genetics at U of Leicester. Genome structural variation.
"Anyone with gumption and a sharp mind will take the measure of two things: what's said and what's done." Views my own.
Functional morphology and physiological ecology of fishes. Postdoc | she/her
Understanding bats through research. Host-Pathogen responses/Comparative Biology/Immunology. opinions my own. A. Prof @ZJE_institute Zhejiang University
Professor of marine biology at James Cook University, Australia - #biodiversity #coralreefs #fish #sharks #stress #physiology #evolution #conservation #climatechange #physioshark #TEDx #LGBTQ #STEM
Professor of Evolutionary Biology at Louisiana State University.
Phylogenomics, Molecular Evolution, Evolutionary History, Statistics, Computational Biology
Indiana and Texas Alum
Professor. Sociologist. NYTimes Opinion Columnist. Books: THICK, LowerEd. Forthcoming: 1)Black Mothering & Daughtering and 2)Mama Bears.
Beliefs: C.R.E.A.M. + the internet ruined everything good + bring back shame.
“I’m just here so I don’t get fined.”
Marine Ecologist, Deep-Sea Explorer, Climate Change Researcher, Science Communicator, Ed @deepseanews
Studying why and how behavior evolves, from mosquitoes to mole-rats | Postdoc/Leon Levy Scholar @Columbia working with Ishmail Abdus-Saboor | PhD @Princeton with Lindy McBride | 麻布/東大 alum 🇯🇵 | yukihaba.github.io
Senior Research Scientist at Field Museum || Lecturer @saic-edu.bsky.social || Evolutionary biologist interested in macroevolution, symbiosis, fungi and geobiology. Posts my own. https://mpnelsen.com/
evolutionary/population/community ecologist
associate professor at NC State
Interests: 🧉🌱🏃🏾♀️🐦⬛🎶🏔️🏜️
www.seemasheth.weebly.com
she/her
views my own
UMich professor who studies resistance and persistence (in plants)