
New #BehindThePaper for our latest paper in @natcomms.nature.com (rdcu.be/e2qMK)
Secret Invasion: Strain Fate Across Microbiomes
Cover illustration by Helena Klein (@illuzation.bsky.social).
communities.springernature.com/posts/secret...
@asanchezlab.bsky.social
CSIC Professor and Principal Investigator of the Quantitative Biology group at IBFG in Salamanca. Previously at Yale EEB, CNB-CSIC. Our group works on building predictive models of biological teams. More information at www.sanchezlaboratory.weebly.com

New #BehindThePaper for our latest paper in @natcomms.nature.com (rdcu.be/e2qMK)
Secret Invasion: Strain Fate Across Microbiomes
Cover illustration by Helena Klein (@illuzation.bsky.social).
communities.springernature.com/posts/secret...
How does population density affect evolutionary trajectory?
Microbes construct their own niche which in turn reshapes their evolution.
Preprint drop from grad student @noahhoupt.bsky.social whose evolution experiments featured blue/white colonies, 1000 generations, a lab move, and much more!
Registration is open for the inaugural GRC conference in the Function of Evolving Systems. Aug 9-14, 2026, Waterville Valley. Truly stellar speaker lineup. Student/postdoc fellowships are available! Please come join us! www.grc.org/function-of-... @joybergelson.bsky.social
29.01.2026 15:03 — 👍 42 🔁 37 💬 1 📌 2 New paper out in @pnas.org, and it made the cover! 👁️
We represent plasmids as circles and mutations as dots, resembling an eye, because in this paper we literally 𝑤𝑎𝑡𝑐ℎ plasmids evolve.
‼️Check Paula’s 🧵 and the paper👇
𝗣𝗹𝗮𝘀𝗺𝗶𝗱 𝗺𝘂𝘁𝗮𝘁𝗶𝗼𝗻 𝗿𝗮𝘁𝗲𝘀 𝘀𝗰𝗮𝗹𝗲 𝘄𝗶𝘁𝗵 𝗰𝗼𝗽𝘆 𝗻𝘂𝗺𝗯𝗲𝗿
www.pnas.org/doi/10.1073/...
Very interesting theoretical paper on BEF and species interactions, presenting an elegant partitioning of species effects on ecosystem function.
www.biorxiv.org/content/10.1...

@jakob-tr.bsky.social @jrpenades.bsky.social et al. 🔥!
www.science.org/doi/10.1126/...
We’re very happy to share the results from the last chapter of my PhD, now out as a preprint on bioRxiv
www.biorxiv.org/content/bior...

Happy to share the latest perspective piece from our lab! www.sciencedirect.com/science/arti...
05.09.2025 14:19 — 👍 7 🔁 4 💬 0 📌 0New year, new conferences! Consider submitting to the symposium on Fitness landscapes and Genotype-phenotype maps, linking computational and experimental approaches (organising with @n-martin.bsky.social; @dbajic.bsky.social invited spreaker) at SMBE (28/6-2/7)! Abstract deadline February 3rd!
09.01.2026 10:37 — 👍 9 🔁 6 💬 0 📌 1Warning though: The job post is in "Spanish" only (in truth, a dialect used exclusively at the BOE), and the instructions for applying could be MUCH more clearly explained on the web, they are difficult to navigate for non-spaniards (even for most of us spaniards too!).
02.01.2026 20:22 — 👍 2 🔁 1 💬 0 📌 0Importantly, there used to exist additional hurdles for scientists with PhDs from non-spanish institutions, but I believe these no longer exist for scientists with PhDs from the European Higher Education Area (hurdles still in place for PhDs from USA, China, Australia, Canada, etc)
02.01.2026 20:22 — 👍 3 🔁 1 💬 2 📌 0Opportunity for scientists considering moving to Spain as tenured scientists! Happy to see openings in microbiology across the country (including at CNB which has incorporated fantastic micro eco/evo researchers lately) and functional & systems biology at IBFG
www.ciencia.gob.es/en/Convocato...

🆕🎙️ El último podcast de 'Ciencia de Cerca' de 2025 llega con un invitado muy especial 🧑💻🦠 Álvaro Sánchez (@asanchezlab.bsky.social) investigador #ERC (@erc.europa.eu) del @ibfg.bsky.social y su novedoso trabajo de "ingeniería" de comunidades microbianas.
Escúchalo en Ivoox👉 go.ivoox.com/rf/164823351
A pleasure and an honour to have received this prestigious grant with my admired colleagues and friends @sanmillan.bsky.social and @asanchezlab.bsky.social . More about AMR under plasmid lens!. My sincere gratitude to @caixaresearch.bsky.social
@microryc.bsky.social @esgem-sg.bsky.social

2/ 🤝 This project is carried out at @cnb-csic.bsky.social is conducted in consortium with @tmcoque.bsky.social (Instituto Ramón y Cajal de Investigación Sanitaria).
20.11.2025 11:03 — 👍 10 🔁 3 💬 0 📌 1
1/ Antibiotic resistance causes ➕ than 1 M deaths each year and understanding why this happens is key to stopping it.
With #HealthResearch support, @sanmillan.bsky.social is investigating how different plasmids (DNA fragments) coexist in bacteria and confer resistance.
https://tinyurl.com/3bysv6cr

Open Call for Expressions of Interest: become a Max Planck Director & redefine what’s possible in science! We offer scientific independence, long-term stable funding & exceptional infrastructure. Lead your own department & open new frontiers of research. Apply by 31 Oct 2025 → www.mpg.de/directors
17.09.2025 15:25 — 👍 39 🔁 37 💬 0 📌 2
Graphic with a blue background and light green dots. The text 'call for applications' appears in green, and 'Ausschreibung' in white. A white arrow on the right points diagonally upward.
Early-career researchers: want to run your own lab? 🌟Max Planck Research Groups offer 6+ years, up to €2.7M in funding, open-topic freedom, team support & tenure-track opportunities. Intrigued? 😃Apply by Oct 14, 2025! www.mpg.de/max-planck-r...
15.09.2025 09:26 — 👍 92 🔁 98 💬 0 📌 4
Antimicrobial Resistance (AMR) could cause 10 million deaths by 2050. At PLAS-FIGHTER, we are working to change this!
🔎 We study how plasmids spread resistance to open up new avenues for future containment and diagnostic strategies.
Find out more 👇
plasmidlab.es/project/
#AntibioticResistance

New paper alert! 🚨
Plasmids promote bacterial evolution through a copy number-driven increase in mutation rate.
We combine theory, simulations, experimental evolution, and bioinformatics to demonstrate that mutation rates scale with plasmid copy number.
Let's dive in! 🧵👇
Do plasmids evolve faster 🐇, slower 🐢, or just like chromosomes 🧬?
In our new paper, we tackled this question using theory, simulations, bioinformatics, and experiments!
👇 Check out all the details in Paula’s thread!
Hint: 🐇 (most of the time)
Check out my first PhD paper on between-species genetic interactions (intergenomic epistasis) with @cbank.bsky.social 🌿↔️🦠✨ We define intergenomic epistasis mathematically and highlight its potential in understanding co-evolution :)
03.04.2025 15:03 — 👍 13 🔁 5 💬 1 📌 0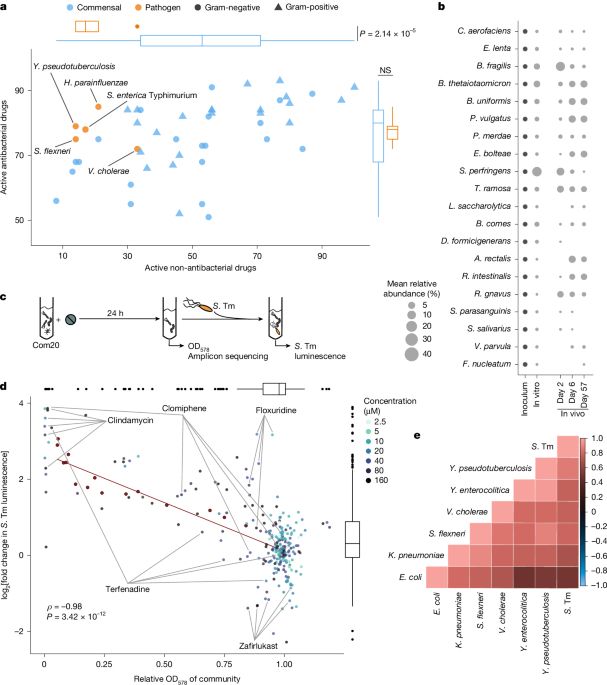
Seeing this published is even better than eating sweets! I am so happy and proud of whole @lisamaierlab.bsky.social, especially Lisa and @jdlcz.bsky.social who walked the funny/exciting/rocky road with me until the very end. Check it out!
#science #microbiome #health
www.nature.com/articles/s41...
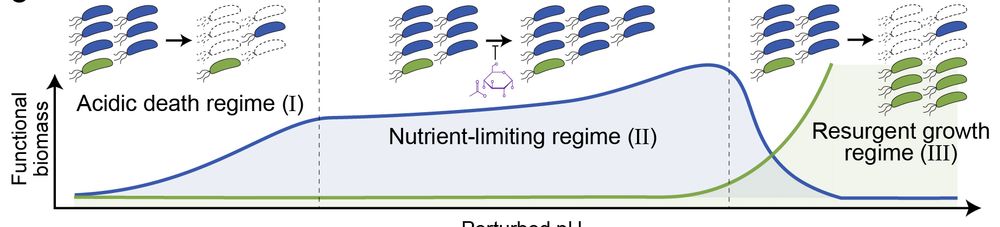
New paper in @nature.com! With @kiseokmicro.bsky.social , Siqi Liu, Kyle Crocker, Jojo Wang, Mikhail Tikhonov & Madhav Mani — a massive dataset and simple model reveal a few conserved regimes that capture how soil microbiome metabolism responds to perturbations. www.nature.com/articles/s41...
17.07.2025 06:36 — 👍 66 🔁 30 💬 2 📌 4This paper by Belda et al makes the case for wine fermentation as a model system in microbial ecology and evolution, one which hits a sweet spot between experimental tractability and rich ecological and evolutionary dynamics.
enviromicro-journals.onlinelibrary.wiley.com/doi/full/10....
This paper by Shraddha Karve discusses how evolutionary novelties emerge in microorganisms, focusing on the different types of novel traits and the role played by the environment.
enviromicro-journals.onlinelibrary.wiley.com/doi/10.1111/...
This piece by Orr, Armitage & Letten provides a nice & clear introduction to modern coexistence theory for microbiologists, discussing how it can be applied to microbial communities & highlighting its promise, limitations & opportunities.
enviromicro-journals.onlinelibrary.wiley.com/doi/10.1111/...

This excellent article by @clsong.bsky.social synthesizes research on the Assembly Graph, a useful conceptual & methodological tool to understand the dynamic assembly of microbial communities and multispecies coexistence.
enviromicro-journals.onlinelibrary.wiley.com/doi/abs/10.1...
This paper by Belda et al makes the case for wine fermentation as a model system in microbial ecology and evolution, one which hits a sweet spot between experimental tractability and rich ecological and evolutionary dynamics.
enviromicro-journals.onlinelibrary.wiley.com/doi/full/10....
This paper by Shraddha Karve discusses how evolutionary novelties emerge in microorganisms, focusing on the different types of novel traits and the role played by the environment.
enviromicro-journals.onlinelibrary.wiley.com/doi/10.1111/...