
Finally, the first public release of #BigVolumeBrowser, so after teasers, you can try it yourself. For details, please check the announcement post (1/2)
forum.image.sc/t/bigvolumeb...
@aafkegros.bsky.social
Check out Microscopy Nodes for handling microscopy data in Blender! Now loading .tif and OME-Zarr with big volume support :) she/her | IPA: ˈafkə ɡrɔs

Finally, the first public release of #BigVolumeBrowser, so after teasers, you can try it yourself. For details, please check the announcement post (1/2)
forum.image.sc/t/bigvolumeb...
Trackastra is part of the brand-new TrackMate v8.
It is the first deep-learning-based ("AI") drop-in replacement for TrackMate's regular LAP-Track algorithm.
Tracks out of the box for many types of datasets (bacteria, fluorescent nuclei, phase contrast cell culture etc), zero parameters to tune 🤖
I agree, is there a way to drop the name from the image mark (or maybe make multiple, one with the name big to the right of the logo?)
29.10.2025 08:45 — 👍 3 🔁 0 💬 1 📌 0
SynapseNet is a deep learning based software tool that automates the segmentation of vesicles, mitochondria, synaptic compartments, and the active zone. This is visualized in three panels. On the left, a section of an electron tomogram with a synaptic compartment densely filled with vesicles, which appear as round structures with dark boundary and light body in the image, is shwon. The top right panel shows the segmentation result of vesicles, visualized by masks with an individual color per vesicle and outlines for the segmented compartment (red) and active zone (blue). The bottom right panels shows a 3D rendering of the segmentation with vesicles shown as yellow spheres.
Are you studying synapses in electron microscopy? Tired of annotating vesicles? We have the tool for you! SynapseNet implements automatic segmentation and analysis of vesicles and other synaptic structures and has now been published:
www.molbiolcell.org/doi/full/10....
Are you rendering all points as emitters? Or do they have some form of occlusion/absorption/scattering?
10.10.2025 13:08 — 👍 1 🔁 0 💬 1 📌 0
Large images have to be broken into tiles both for training and inference with neural networks. The tile predictions then need to be merged to produce the final volume prediction.
Segment large images without tiling artifacts: sharing our work that should have been presented at ICCV in 2 weeks - the brilliant first author Elena can’t go because of visa issues.
The paper: arxiv.org/abs/2503.19545 1/🧵
Ah right, I have also seen some of the napari solution! I think this in general makes a lot of sense, and I'm still looking into how to make these kinds of volume interactions as easy in Blender as they are in the more scientific software, such as BigTrace and Napari 😅😋
24.09.2025 11:29 — 👍 1 🔁 0 💬 1 📌 0This is separate from Blender, built on imglib2 in java (same framework as bigDataViewer, so also related to bigWarp), and for bigtrace all implemented by @ekatrukha.bsky.social
I think the smart thing it does for annotation is to extract the brightest point under the mouse as targeted point
This is the cool part of BigTrace, where this also has extensive semimanual and manual modes, while seeing your data in 3D. So this way, you can use the same engine to delete and fix traces.
Additionally i still want to make some automatic flags based on time info, but that will be more custom.
Automatic filament tracing in 3D data is now only a few clicks (or one macro) away! In the amazing #BigTrace FIJI plugin by @ekatrukha.bsky.social 🤩
My workflow will be to autotrace and then manually correct the autotraces, reducing the time for our tracing significantly!
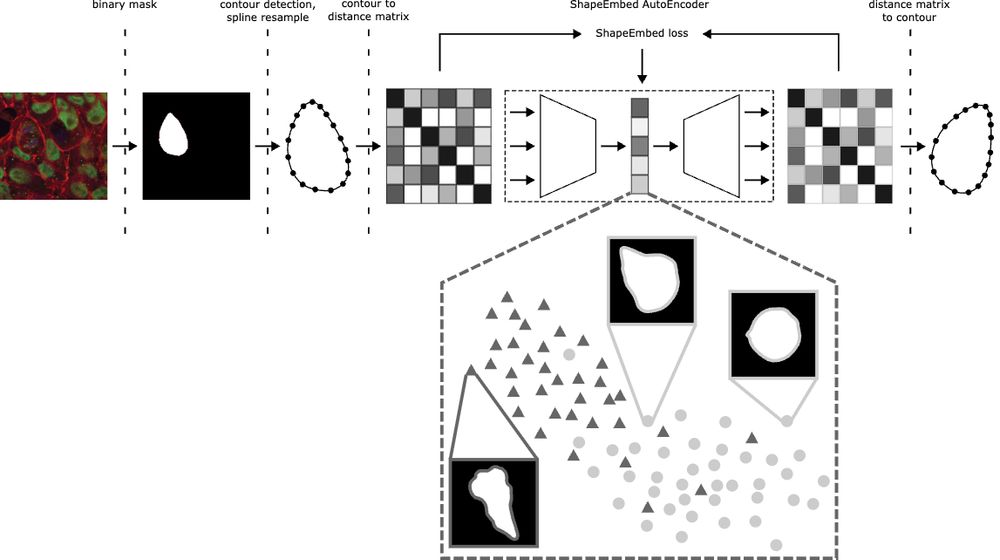
Happy to share that ShapeEmbed has been accepted at @neuripsconf.bsky.social 🎉 SE is self-supervised framework to encode 2D contours from microscopy & natural images into a latent representation invariant to translation, scaling, rotation, reflection & point indexing
📄 arxiv.org/pdf/2507.01009 (1/N)
A cell finding its way through the matrix, imaged with @joycemeiri.bsky.social on LLS.
19.09.2025 10:38 — 👍 34 🔁 13 💬 1 📌 1Amazing to see real-world medical applications of blender volume handling! 🤩🤩🤩
19.09.2025 12:44 — 👍 6 🔁 0 💬 1 📌 0
I gave a talk at Blender Conference yesterday, showing how biological volumes work, and how to visualize them in Blender 😋 youtu.be/WPajSWX730o?... #bcon25
19.09.2025 07:35 — 👍 57 🔁 15 💬 3 📌 2I think it can be very useful to give a 'diorama' feel to a render and highlight certain complexity! But probably mostly for artistic purposes
27.08.2025 17:00 — 👍 1 🔁 0 💬 1 📌 0
How do you quantify cell structures which are deformed between different observations? Our preprint shows how to perform Simultaneous QUAntification of Structure and Structural Heterogeneity (SQUASSH) on 3D fluorescence microscopy data. (1/5)
www.biorxiv.org/content/10.1...
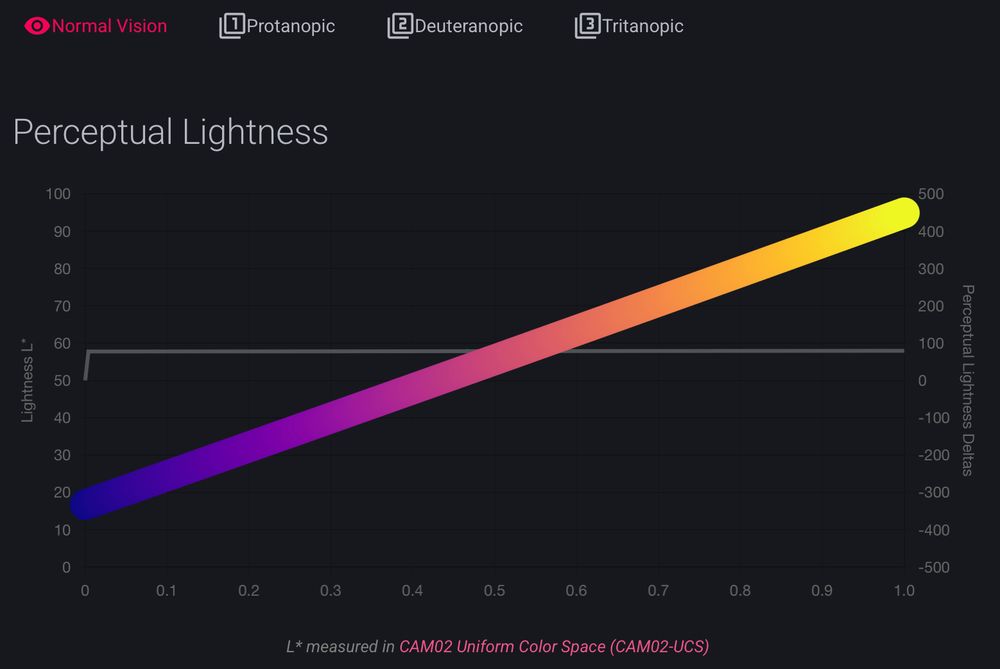
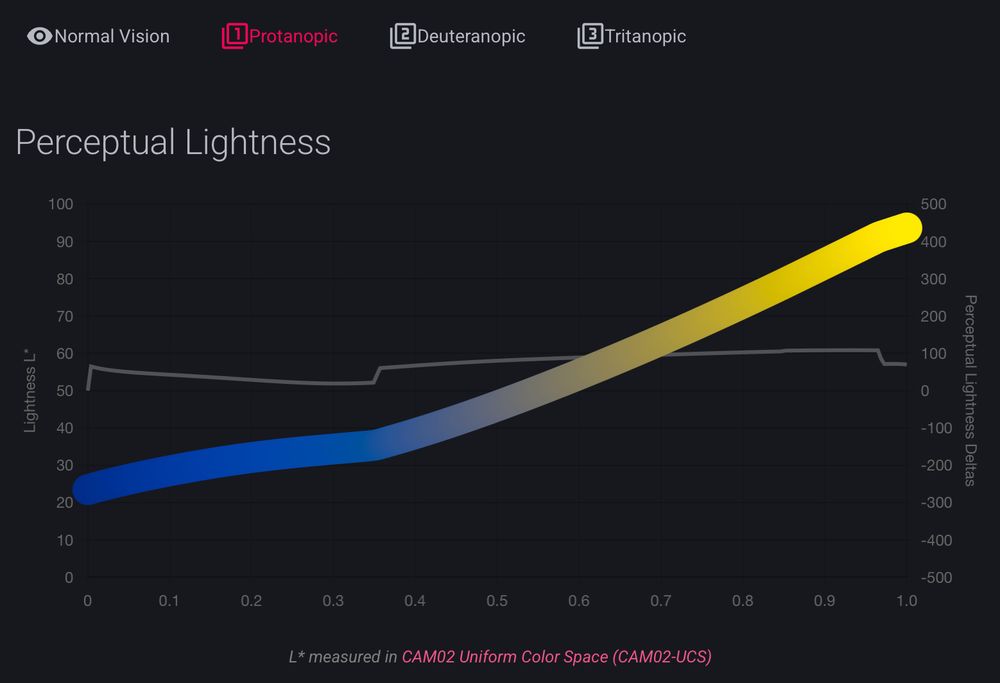
it was already fun to look around the documentation for cmap (cmap-docs.readthedocs.io)
but @grosoane.bsky.social just added a very cool feature to toggle between color vision deficiencies. Watch your perceptually uniform color maps gain some kinks 😂
as a mild deutan myself, I appreciate it! 🙏
Happy to officially introduce FilaBuster - a strategy for rapid, light-mediated intermediate filament disassembly. Compatible with multiple IF types, modular in design, and precise enough to induce localized filament disassembly in live cells.
www.biorxiv.org/content/10.1...
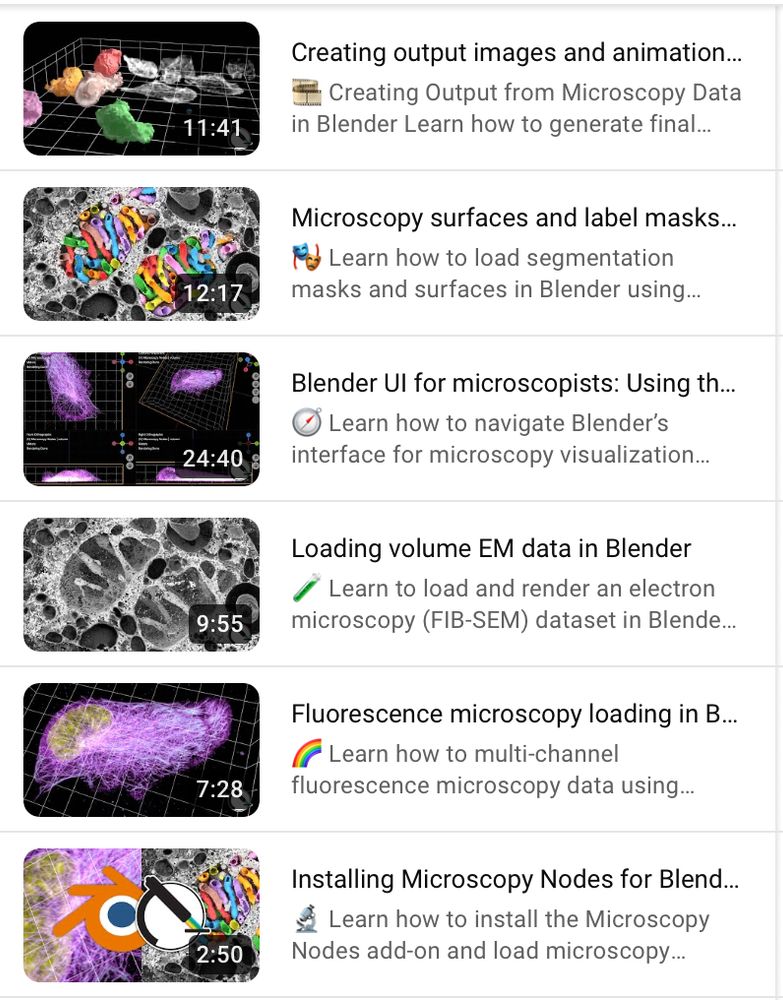
This is accompanied by a new set of tutorials showing exactly how you can start making beautiful microscopy visualizations in @blender.org!
Following along is also easy with the example OME-Zarr @openmicroscopy.org datasets 😄
Find the tutorials here: www.youtube.com/playlist?lis...
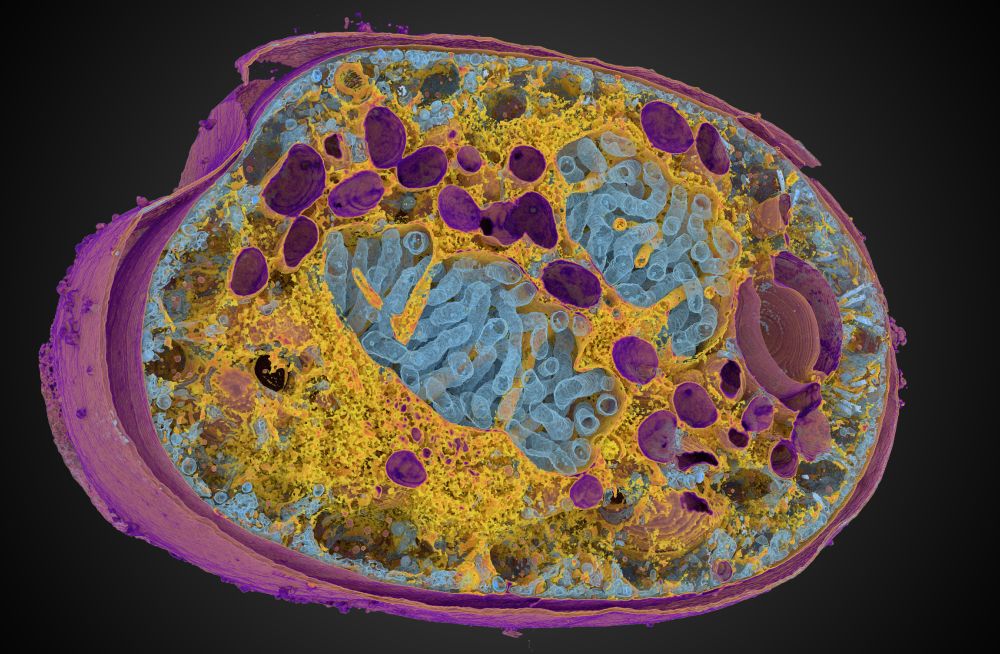
In the context of our @reviewcommons.org revision process, I'm happy to announce Microscopy Nodes v2.2.0!
This packs lots of new fun features, including new color management 🌈, clearer transparency handling 🫥, custom default settings 🔧 and more!
Preprint at doi.org/10.1101/2025...
New stable ilastik version 1.4.1 at www.ilastik.org/download!
Release Highlights:
- Support for OME-zarr (v0.4)
- New workflow - Trainable Domain Adaptation
- New object features based on Spherical Harmonics Feature, contributed by @grosoane.bsky.social.
- Native support for Apple Silicon
So excited to be a collaborator on this! We used @grosoane.bsky.social #microscopynodes to import tomo data to Blender before passing to Sensu for the beautiful animation.
03.04.2025 16:50 — 👍 7 🔁 1 💬 0 📌 0
It would only be appropriate to post about the #geometrynodes #microtubule here too. For a full introduction, I have prepared the world's least professional youtube video.
www.youtube.com/watch?v=AYUy...
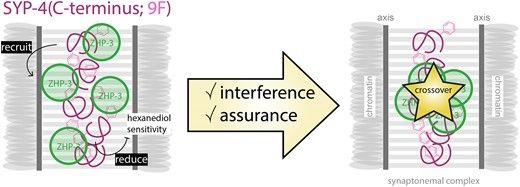
My PhD work in the Köhler lab showing an involvement of a synaptonemal complex component in crossover regulation independently of synapsis is now published in NAR! 🎊 #meiosis #crossovers #C.elegans 🧬
Check it out here: academic.oup.com/nar/article/...
Together with Christian Tischer, 3D rendering of multi-res microscopy data was added to MoBIE FIJI plugin using #BigVolumeViewer.
Check any remote datasets (like @dudinlab.bsky.social maps of protists using ExM) in the highest quality.
For full details read forum.image.sc/t/mobie-plus...
10 - Featured extension: Mastodon-Blender
Offers a bridge from Mastodon to Blender to create high quality 3D rendering of lineages and cell tracks over time:
(Tracking data of phallusia mammillata embryogenesis by Guignard et a. (2020). doi.org/10.1126/scie...)
And it also works in 2D 😁 Here we extract a circular projection instead of a spherical, and apply a 1D Fourier transform.
Here we can see how the channel distributions all align in the projection after we optically activate Rac1 in one part of the cell. 💡
With @jpassmore.bsky.social , Lukas Kapitein
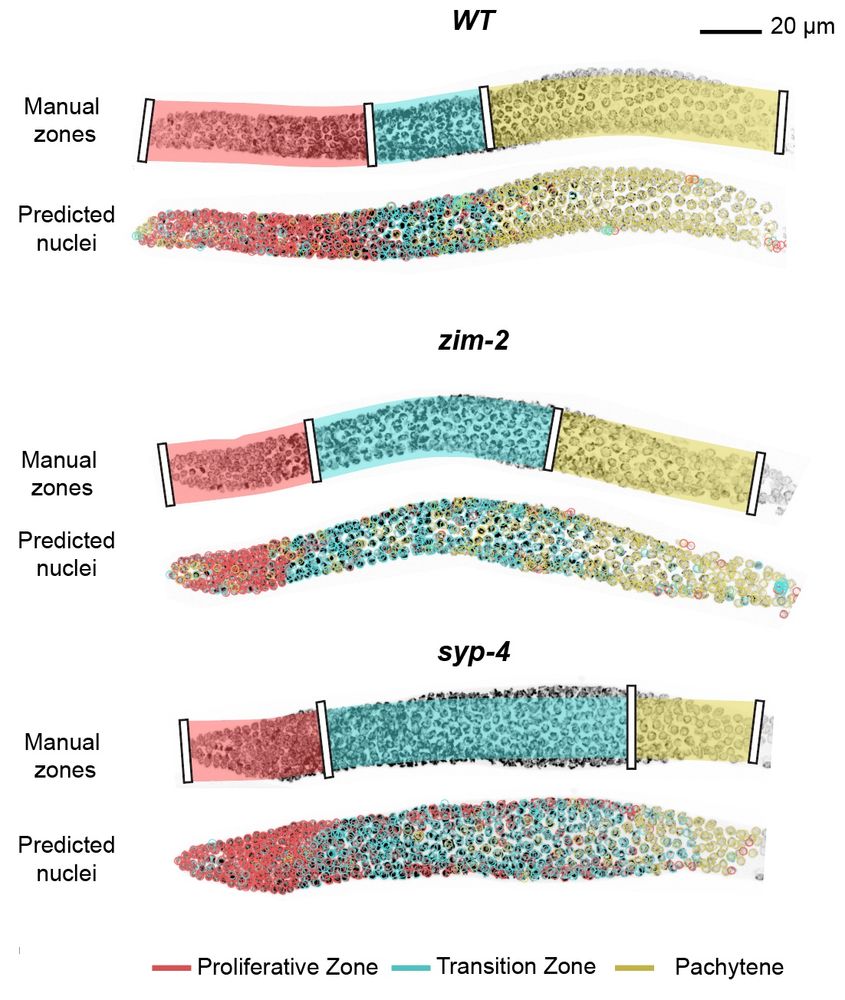
With the integration into the accessible toolkit ilastik (@ilastik-team.bsky.social) you can very easily include this in a quickly and interactively retrainable classification algorithm.
We show this for classification of meiotic zones in C. elegans :) 🪱
For the nerds there's also a Python API 😉
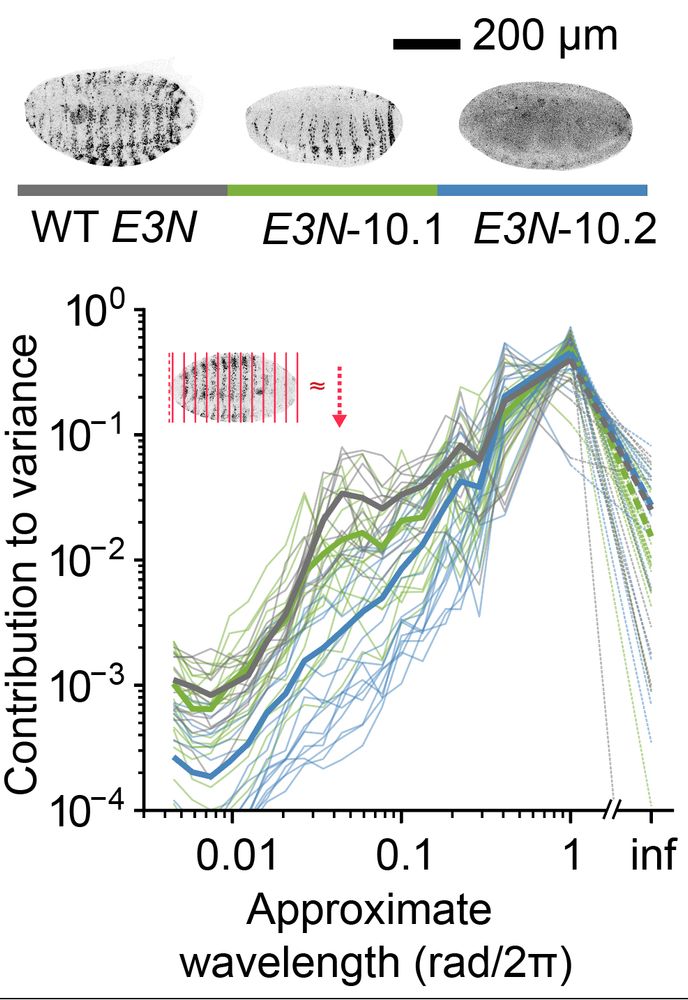
Quantifying distribution of intensity can be very helpful in many biological systems. For example, in D. melanogaster data, we extract a peak for the level of embryonic patterning in development. 🪰
In collaboration with
@ottilie.bsky.social, @timothyfuqua.bsky.social, @justinmcrocker.bsky.social
We then use spherical harmonics decomposition to quantify the variance in the spherical signal per angular wavelength.
This gives us a metric that shows how detailed the features in the spherical map are, giving a rotation-invariant quantification of distribution.