Paraviewer creates easy-to-use review websites from Paraphase output. Try these demos!
WGS data: pacificbiosciences.github.io/ParaviewerWG...
PureTarget data: pacificbiosciences.github.io/ParaviewerPT...
Paraviewer creates easy-to-use review websites from Paraphase output. Try these demos!
WGS data: pacificbiosciences.github.io/ParaviewerWG...
PureTarget data: pacificbiosciences.github.io/ParaviewerPT...

My new tool Paraviewer is now available for use at github.com/PacificBiosc...! If you use Paraphase, try this new next-step tool - it automates and greatly simplifies variant visualization from Paraphase variant calling. If you're at #ASHG2025, visit me today at poster 4109W. #pacbio
15.10.2025 14:58 — 👍 7 🔁 5 💬 1 📌 0
Attending #ASHG25? Visit #PacBio at booth 919!
Connect with our team, explore live demos of #Vega, and discover the latest in long-read sequencing. While you’re there, enter our daily Labubu drawing!
See the full program here: bit.ly/4nmBnfn
#ASHG #HumanGenomics #ASHG2025

Tool (with demos) available at github.com/PacificBiosc...
09.10.2025 19:01 — 👍 0 🔁 0 💬 0 📌 0
Here's an example from the paper: a complex rearrangement shown with svtopo (top), IGV (middle), and Ribbon (bottom):
09.10.2025 19:00 — 👍 0 🔁 0 💬 0 📌 0
My complex variant visualization tool SVTopo is now officially published in BMC Genomics! link.springer.com/article/10.1.... This tool allows HiFi users to view complex germline structural variation in intuitive and informative plots.
09.10.2025 19:00 — 👍 6 🔁 7 💬 2 📌 0
I'm excited to share our pre-print about a new variant benchmarking tool we've been working on for the past few months!
Aardvark: Sifting through differences in a mound of variants
GitHub: github.com/PacificBiosc...
Some highlights in this thread:
1/N
Install SVTopo from conda, or go straight to the source at github.com/PacificBiosc.... If you break it, let me know! The best way is by creating a github issue on the repo.
22.04.2025 14:25 — 👍 1 🔁 0 💬 0 📌 0In addition to making these handy-dandy images, SVTopo creates a serverless table browser to sort, filter, and view the results. The one for these 7 samples is live at pacificbiosciences.github.io/SVTopo/. Try it out!
22.04.2025 14:25 — 👍 1 🔁 0 💬 1 📌 0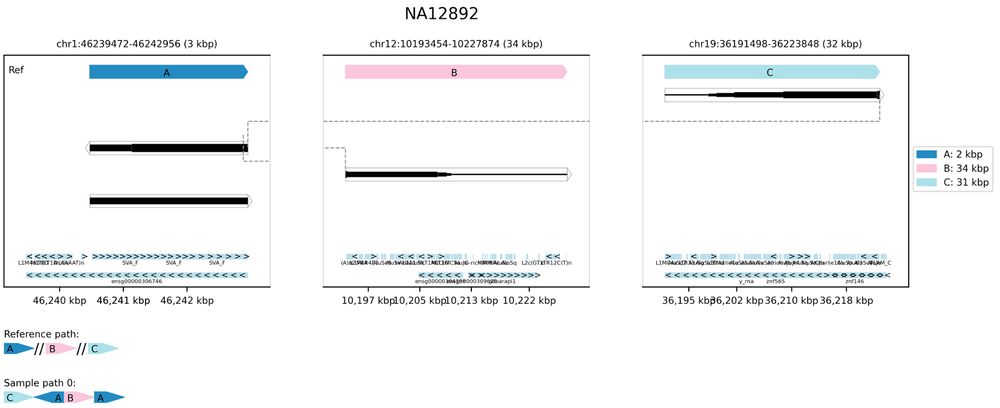
This example might look at first glance like a bad representation of a translocation, but the SVA annotation is a giveaway - reads from chr12 and chr19 aligned to a random SVA retrotransposon rather than a real connection between three chromosomes.
22.04.2025 14:24 — 👍 1 🔁 0 💬 1 📌 0Alignment challenges mean that the visualization doesn’t always work, of course. 125 images from these 7 genomes had alignment artifacts that made them useless for variant interpretation. Many of these appear to be useful in another way, though – identification of false positives.
22.04.2025 14:23 — 👍 1 🔁 0 💬 1 📌 0
We looked at SVTopo images for 7 unrelated samples (HG002 and 6 from the platinum pedigrees cohort: 10.1101/2024.10.02.616333) and observed 142 unique complex SVs that we put in 11 categories (fig2).
22.04.2025 14:23 — 👍 1 🔁 0 💬 1 📌 0
This example is hard to understand from e.g. IGV/Ribbon (see Fig1) but pretty simple in SVTopo: 4 blocks deleted (B,D,F,H), 2 inverted (E,G), 1 re-ordered (C)
22.04.2025 14:22 — 👍 2 🔁 1 💬 1 📌 0Although there are some excellent genome visualization tools out there, none of them are great at representing the complex structural variation that long reads uniquely identify. SVTopo is for this. Shows alignments in new genomic blocks and represents those blocks relative to the reference genome.
22.04.2025 14:22 — 👍 1 🔁 0 💬 1 📌 0
I just released a new preprint! The manuscript describes SVTopo, a software tool that enhances visualization of complex SVs using HiFi data: www.biorxiv.org/content/10.1.... Here’s a summary of the results:
22.04.2025 14:21 — 👍 13 🔁 8 💬 1 📌 1
Great to see that sawfish, our new HiFi SV caller, is accepted for publication in Bioinformatics! Sawfish emphasizes local haplotype modeling to improve SV representation and genotyping in both single and joint-sample analysis. Advance-access article now available: (1/n)
doi.org/10.1093/bioi...
Interesting comparison between long-read sequencing techs for metagenomics: papers.ssrn.com/sol3/papers....
22.01.2025 20:32 — 👍 0 🔁 0 💬 0 📌 0
“StarPhase: Comprehensive Phase-Aware Pharmacogenomic Diplotyper for Long-Read Sequencing Data” is now on biorxiv! In this work, we explore the use of long-read sequencing (#PacBio #HiFi) for #pharmacogenomics #PGx. 1/N
Pre-print: doi.org/10.1101/2024...
Repo: github.com/PacificBiosc...
Another very insightful paper on #methylation and long-read sequencing by @gangfang.bsky.social, describing some important but frequently overlooked caveats.
Gang provides a fantastic summary in his post, but here's my take too😅:
If you were interested but missed it: www.pacb.com/wp-content/u...


#ASHG2024 was a blast. I had great discussions with friends and #pacbio collaborators, presented a poster on my new tool SVTopo, welcomed #PacBioVega to the world, and realized than I'm a bigger fan of One Republic than I knew before. Excited for next time!
13.11.2024 18:59 — 👍 11 🔁 1 💬 1 📌 0
It's official, #PacBio has launched a new benchtop sequencer!
Quick summary of the new Vega system:
- instrument = $169k
- consumables = $1100 per run
- output = 60 Gbp, 24 hr run time
This is the #HiFi sequencer #microbiology labs have been asking for.
www.pacb.com/press_releas...
Glad to see the interest! Associating individual alignments with VCF breakend coordinates can be challenging, which is why the recommended workflow uses sawfish (we added a sawfish read output to make it work) but svtopo can also run with just the HiFi bam for samples without sawfish calls
30.10.2024 15:57 — 👍 1 🔁 0 💬 0 📌 0