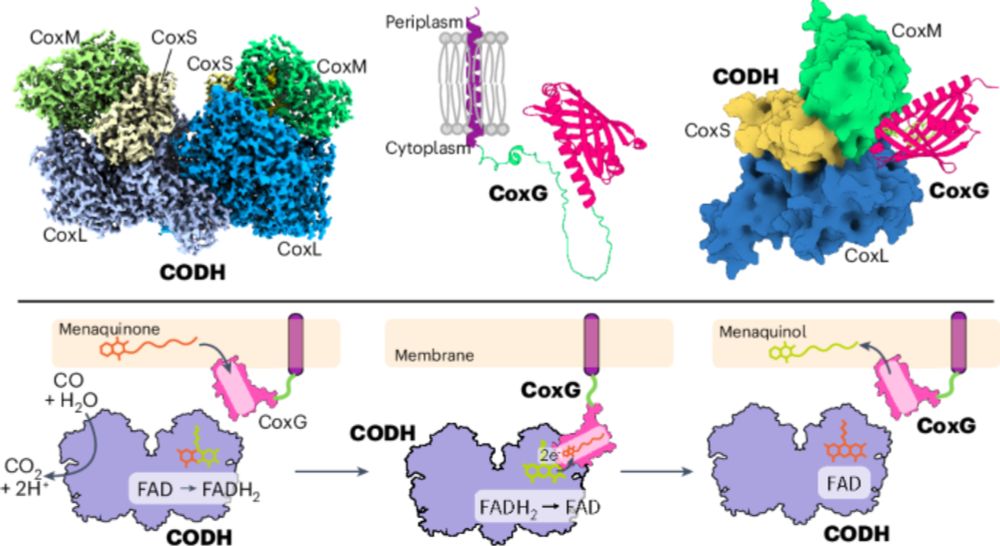
How can AI and #cryoEM help save some of the biggest babies on the planet? 🐘🔬
Our new preprint identifies Elephant Herpesvirus gH/gL/gO as a receptor-binding complex and a promising vaccine candidate, now being tested in young elephants across European zoos! 💉
www.biorxiv.org/content/10.6...
04.12.2025 08:43 — 👍 35 🔁 8 💬 1 📌 2
This one is a bit of a departure from the usual and definitely a work in progress!
We found that by using ab initio reconstruction at very high res, in very small steps, we could crack some small structures that had eluded us - e.g. 39kDa iPKAc (EMPIAR-10252), below.
Read on for details... 1/x
13.09.2025 00:04 — 👍 266 🔁 82 💬 20 📌 14
Was an absolute pleasure to write with @cyntiat.bsky.social, Janik, @rhyswg.bsky.social and @knottrna.bsky.social 😊
02.09.2025 04:08 — 👍 1 🔁 0 💬 0 📌 0

Code to complex: AI-driven de novo binder design
The application of artificial intelligence to structural biology has transformed protein design from a conceptual challenge into a practical approach …
Quite proud of this one. Our review on AI-designed de novo binders out now in Structure. We give a brief history, outline the current ecosystem, highlight some standout applications and discuss the need for regulatory discussions. 🤖🧪🔬
02.09.2025 04:06 — 👍 19 🔁 5 💬 1 📌 0
LinkedIn
This link will take you to a page that’s not on LinkedIn
Did you know that some bacteria can generate ATP, the energy currency of life, literally out of thin air? 🌬⚡️
In PNAS today, we show how bacteria can do this using just three enzymes. Honoured to contribute to this study with Christoph von Ballmoos, Chris Greening and Gregory Cook
lnkd.in/gxm8SXxF
25.07.2025 05:59 — 👍 6 🔁 1 💬 0 📌 0
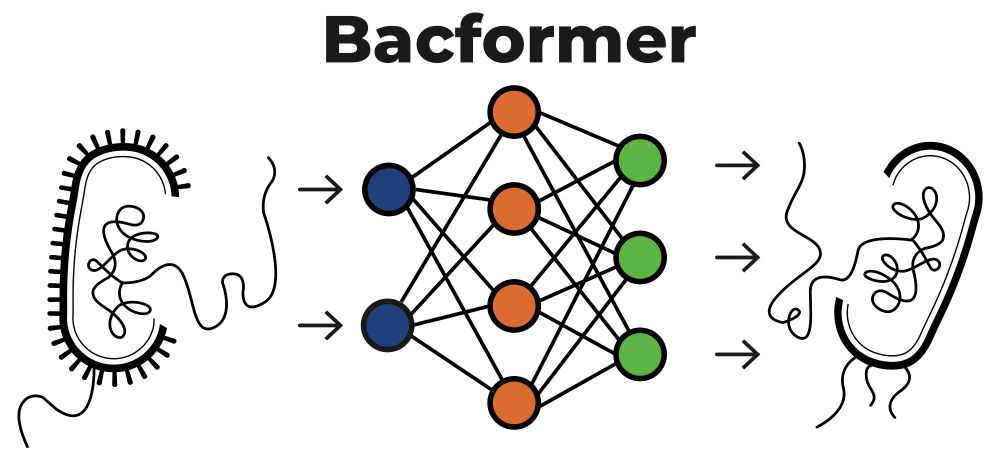
💥 Excited to introduce Bacformer 🦠 - the first foundation model for bacterial genomics. Bacformer represents genomes as sequences of ordered proteins, learning the “grammar” of how genes are arranged, interact and evolve.
Preprint 📝: biorxiv.org/content/10.1...
🧵 1/n
21.07.2025 09:55 — 👍 91 🔁 59 💬 3 📌 2

WEBINAR: Leveraging deep learning to design custom protein-binding proteins
Be inspired by case studies that show you how deep learning methods are speeding up the process of designing custom proteins
Last reminder- Dr Rhys Grinter webinar @rhyswg.bsky.social is on in just a couple of hours- click through to obtain the link. #proteindesign #structbiol
You can sign up for future event information and reminders here:
australian-structural-biology-computing.github.io/website/
🧶🧬
14.07.2025 23:32 — 👍 6 🔁 5 💬 1 📌 0
Thank you!!
10.07.2025 09:01 — 👍 0 🔁 0 💬 0 📌 0
Thank you so much for your help!!
10.07.2025 02:29 — 👍 1 🔁 0 💬 0 📌 0
New to nature proteins are the future! Proud to have contributed to this groundbreaking work! 🏴☠️
10.07.2025 00:03 — 👍 4 🔁 1 💬 0 📌 0
Proud to share some of the work from my PhD is now out in Nature Communications! Very grateful to all coauthors who helped make this happen, especially my supervisors @rhyswg.bsky.social and @knottrna.bsky.social for their support. 🩸 🦠 🔬
10.07.2025 02:08 — 👍 16 🔁 3 💬 2 📌 0
Many thanks to @rhyswg.bsky.social and @greening.bsky.social for supporting my application to attend! 😊
02.06.2025 23:15 — 👍 2 🔁 1 💬 0 📌 0

Australia is in an extinction crisis – why isn’t it an issue at this election?
Some of the country’s most loved native species, including the koala and the hairy-nosed wombat, are on the brink. Is this their last chance at survival?
96% of Australians want more done to protect nature. Meanwhile, just 0.06% of the budget went to on-ground biodiversity programs.
We’re losing species we love & major parties are sleepwalking through it.
Australians care. It’s time politicians did too.
www.theguardian.com/australia-ne...
06.04.2025 20:54 — 👍 292 🔁 100 💬 9 📌 4
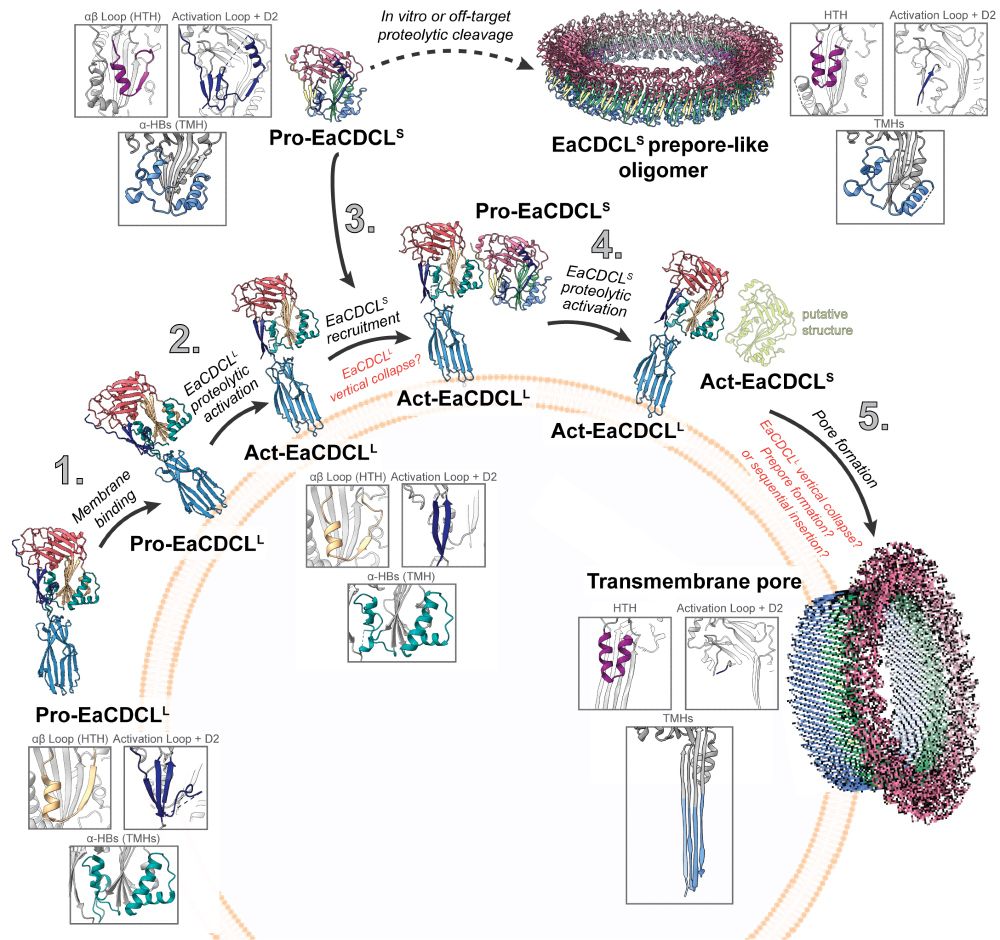
Excited to share that our work on a family of pore-forming proteins is now live in Science Advances! We show structural snapshots across the entire pore-forming pathway for a cholesterol-dependent cytolysin-like (CDCL) bicomponent system.
30.03.2025 10:50 — 👍 32 🔁 9 💬 3 📌 0
Job Search
We are hiring! The Formosa Group is hiring a Research Officer to join our program on #Mitochondria, #Metabolism & #MitochondrialDisease at the Monash BDI!
💡If you're passionate about mitochondrial research or know someone who is, please share! Apply now: careers.pageuppeople.com/513/cw/en/jo...
30.01.2025 06:53 — 👍 5 🔁 4 💬 0 📌 1
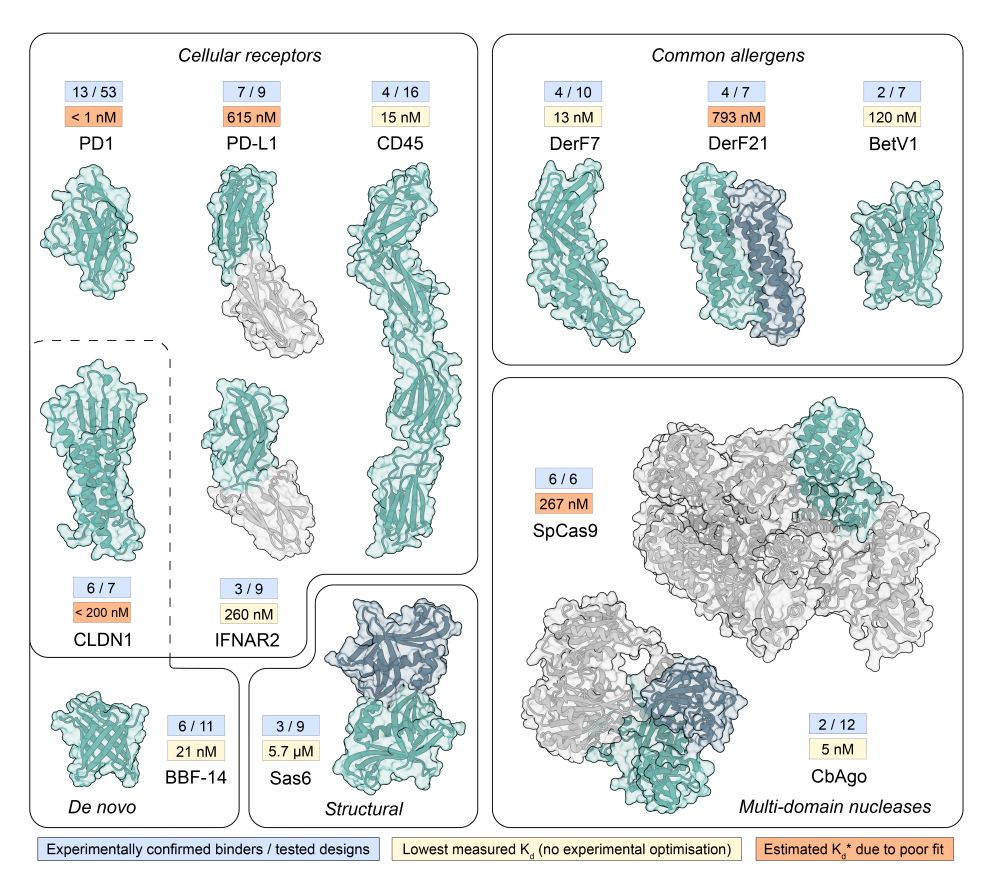
We updated our BindCraft preprint with lots of new exciting results! We release all our binder sequences and models, include more in silico analysis, novel design targets, and present AAV retargeting to specific cell types using de novo binders!
www.biorxiv.org/content/10.1...
08.12.2024 08:26 — 👍 128 🔁 31 💬 1 📌 3
Can I be added please 🙏🏼
11.12.2024 23:44 — 👍 0 🔁 0 💬 0 📌 0
Inhibiting heme-piracy by pathogenic Escherichia coli using de novo-designed proteins https://www.biorxiv.org/content/10.1101/2024.12.05.626953v1
06.12.2024 12:45 — 👍 1 🔁 1 💬 0 📌 0
PhD student at Uni of Cambridge, UK 🔬 | AI for protein design & engineering 🧬 | biotech & environmental applications 🌱 | enzymes 🏗️
🇦🇹🇨🇭
Asst. Prof. Uni Groningen 🇳🇱
Comp & Exp Biochemist, Protein Engineer, 'Would-be designer' (F. Arnold) | SynBio | HT Screens & Selections | Nucleic Acid Enzymes | Biocatalysis | Rstats & Datavis
https://www.fuerstlab.com
https://orcid.org/0000-0001-7720-9
Some kinda compu-structural-biologi-informatician
Bioinformatics, protein modeling, cryoEM, drug screening, function prediction. Daisuke Kihara, professor of Biol/CS, Purdue U. https://kiharalab.org/ YouTube: http://alturl.com/gxvah
Microbiologist. Lead developer of #Kaptive + bonafide #Klebsiella nerd. Post-doc in the Wyres Lab @AlfredMonash_ID.
Structural Biologist 🧬 @WEHI ⚕️ | Protein and Drug Design | Biophysics 🔬 | AI | Apoptosis | Epigenetics | Coffee ☕ | Naarm/Melbourne 🇦🇺
Discover the #ImmuneSystem with #StructuralBiology. We 💗 powerful #Tcells and #viruses. From one of my student “the GRAS is greener in our lab”. #scienceIsFun
Assoc. Prof. at the School of Science, University of Wollongong, Australia. #CryoEM Research Group Leader at Molecular Horizons Research Institute. Biology BS, Biotechnology MS, PhD in Biochem. & Mol. Biol., Post-doc in Molecular Imaging (#UNC, #NIH).
microbiologist, McMaster U 🇨🇦 • type IV pili, phages, biofilms, antibiotic resistance • equestrian🐴 • cancer survivor🎗️
Researcher in situ structural biology using CryoET (Melb, Australia). Father, Greyhound companion, coffee enthusiast, and willing but not able golfer.
Scientist at St Jude using structural biology to try and understand how things work - views are my own not my institutions
Structural biologist interested in all things antibody and viruses. 🏳️🌈
bajiclab.org
Cryo-electron microscopist @ WEHI
Postdoc @Komander lab, WEHI, Melbourne.
Mitochondrial biochemist interested in ubiquitin 🤓.
Scientist (mostly #cryoEM) | Proud father | Avid sports fan moonlighting as an international touch football referee | Rock singer trapped in a tone deaf body
The Australian Society for Biochemistry and Molecular Biology (ASBMB) aims to advance Biochemistry and Molecular Biology in Australia.
This year we are hosting the bi-annual ASBMB conference at the University of Queensland (29th September - 1st October)
Joint Head of Structural Studies at @mrclmb.bsky.social. Develops & uses #cryoEM to study amyloids in neurodegeneration. #tau, #alphasynuclein, #opensoftware, #RELION. All opinions my own.
Senior Research Fellow and Group Leader at @IMBatUQ
Interested in the structural and functional biology of transporters 🇦🇺🔬
Assistant Professor of Biochemistry at the University of Toronto | Virology, structural biology, chemical biology | He/Him. 🇨🇦🏳️🌈👨🔬