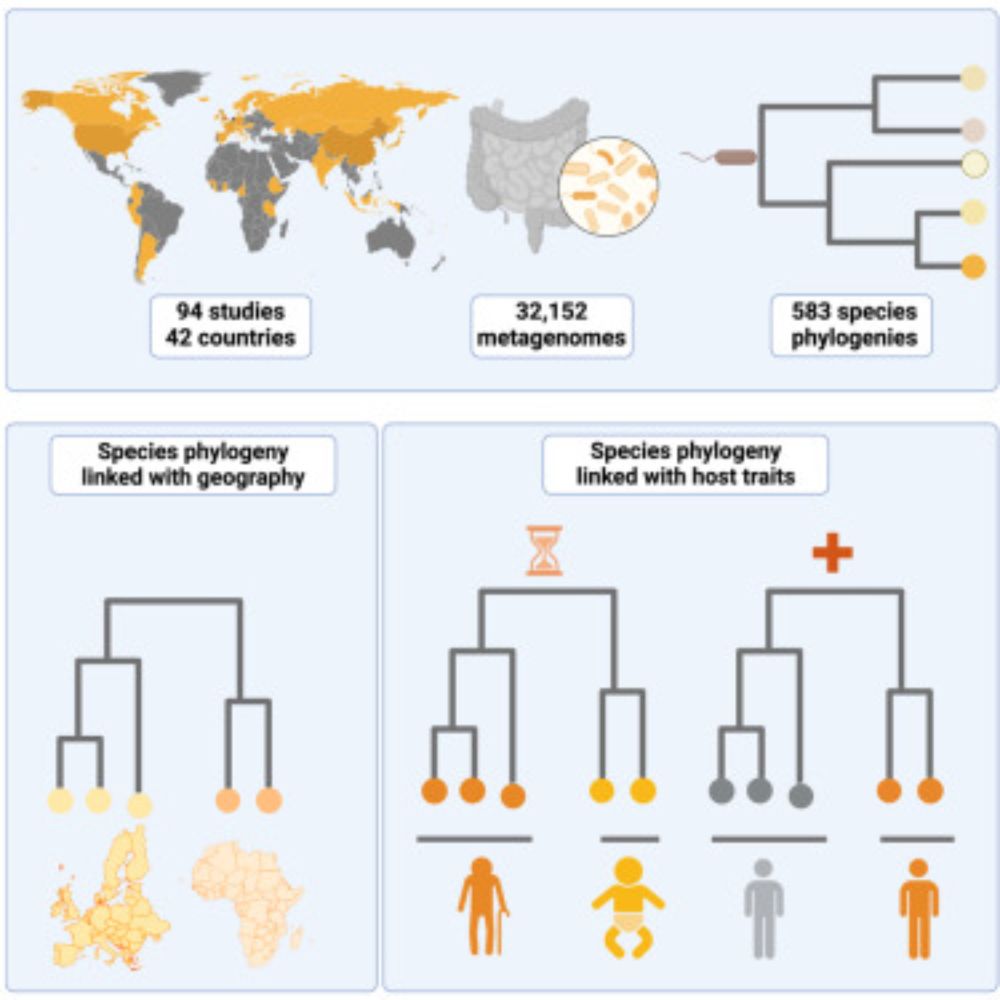
gutSMASH 2.0: Extended Identification of Primary Metabolic Gene Clusters From the Human Gut Microbiota
Microbiota-derived metabolites serve as key messengers mediating host–microbe and microbe–microbe interactions, often through specialized primary meta…
Now available online: the new 2.0 version of gutSMASH, with capabilities to detect 12 new types of catabolic gene clusters relevant to gut microbiome ecology, as well as predictions of their regulation through transcription factor binding site detection. www.sciencedirect.com/science/arti...
13.12.2025 07:41 — 👍 35 🔁 18 💬 2 📌 0

Diagram illustrating "feed the enemy's enemy": ampicillin inhibits Pseudomonas aeruginosa. Citrobacter freundii acidifies the environment, further inhibiting P. aeruginosa.
New preprint from our lab www.biorxiv.org/content/10.1...! Andrea Dos Santos and Clément Vulin combine experiments and models showing how adding glucose can strengthen negative interactions between microbial species. This can be used in tandem with antibiotic treatment to inhibit pathogens!
02.12.2025 13:12 — 👍 62 🔁 33 💬 3 📌 0

🚨Preprint alert - this is a big one! We transfer the revolutionary power of TnSeq to bacteriophages.
Our HIDEN-SEQ links the "dark matter" genes of your favorite phage to any selectable phenotype, guiding the path from fun observations to molecular mechanisms.
A thread 1/8
20.11.2025 20:39 — 👍 210 🔁 90 💬 11 📌 5
Super excited that the bulk of my PhD work is now preprinted! Here we used whole-community competition, or coalescence, experiments to quantify selection acting on genetically diverged strains within larger communities. (1/n)
www.biorxiv.org/content/10.1...
11.11.2025 17:14 — 👍 102 🔁 48 💬 3 📌 2
New paper out 🎆 When an antibiotic-resistant E. coli strain lands in our gut microbiome, whether it will get established or not depends on the ecological context. We studied how other microbes, nutrients, and antibiotic exposure shape its fate.👇
09.11.2025 21:34 — 👍 3 🔁 1 💬 0 📌 0

Phage diversity mirrors bacterial strain diversity in the honey bee gut microbiota - Nature Communications
Authors analyse paired viral and bacterial shotgun metagenomics data from individual honeybee guts, revealing modular, nested phage–bacteria networks, with viral diversity mirroring bacterial strain c...
🐝🦠 New paper: rdcu.be/eOf7A
Phages may drive microbial diversity, yet we often don’t even know how phages & bacteria correlate in nature. Our new study tackles this in the honeybee gut, thanks to the great work of PhD student @malickndiaye.bsky.social at @dmf-unil.bsky.social @fbm-unil.bsky.social
04.11.2025 20:36 — 👍 56 🔁 23 💬 1 📌 3
Our high-precision metagenomic strain caller, PHLAME, is now published in Cell Reports!! www.cell.com/cell-reports...
PHLAME works on tough sample types -- including those with coexisting strains of a species and low depth.
15.08.2025 16:33 — 👍 47 🔁 26 💬 2 📌 0
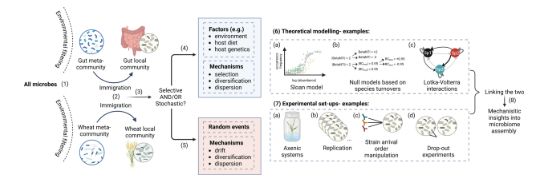
Mechanisms of microbiome assembly and approaches to uncover the ecological forces driving it.
Deriving ecological models solely from observational data limits our ability to understand mechanisms driving microbiome assembly. This #mSystems articles explores how experimental approaches can address these challenges. asm.social/2wj
30.07.2025 15:26 — 👍 11 🔁 8 💬 1 📌 0
Our paper demonstrating that within-species warfare interactions are ecologically important on human skin is now published in Nature Micro! www.nature.com/articles/s41...
30.06.2025 12:26 — 👍 222 🔁 97 💬 10 📌 3
This is such a cool read on the fascinating topic of nutrient competition in microbiomes😍 I couldn t agree more with «A key challenge in understanding microbiomes is that the species composition often differs among individuals, which can thwart generalization. »
13.06.2025 20:19 — 👍 1 🔁 0 💬 0 📌 0
1/6 Excited to share our review 🚀
🧬 CRISPR–Cas therapies targeting bacteria!
#MicroSky
15.05.2025 06:53 — 👍 63 🔁 35 💬 1 📌 3
PhD Candidate at Biozentrum, University of Basel
Microbiology PhD student @ Uni of Birmingham, specialising in bacteriophage, AMR and the gut microbiome. Award winning disability campaigner 🏳️🌈 #ActuallyAutistic, #ADHD, she/her.
I play basketball & videogames and do research. Not necessarily in that order. #RyC Group Leader at IPLA-CSIC.
https://msb-lab.github.io/en
PhD student at Bangor University |
Host-associated microbiomes |
Microbial ecologist |
🇲🇽🏴🏳️🌈
Postdoc - high-throughput experimental evolution - AMR @ University of Oxford
Interested in microbial ecology & evolution. Views are only my own. (he/him)
🎓: https://scholar.google.com/citations?hl=en&user=OBPpZq4AAAAJ&view_op=list_works&sortby=pubdate
👨💻: https://github.com/raufs
PhD student at ETH Zurich
Pathogen Ecology Group (Alex R. Hall)
Nasal microbiome & Staphylococcus aureus 🧫
Postdoc @ Duchossois Family Institute @ UChicago Pamer Lab. PhD Alumni @ IGCiencia #gut #microbiome #consortia #diet #machinelearning #Klebsiella
Synthetic Biology with a Southern European flavour for a new partnership with the Living World https://vdl-lab.com/ http://seva-plasmids.com/
assistant professor at georgetown | polymicrobial interactions | bacterial physiology | science educator | views my own
www.zarrellalab.org
Enjoying the little things like antibodies, ion channels and pip installs - technical biologist building tools and studying diseases ❤️🐢
Asst. Prof. at University of Geneva
Evolutionary cell biology & multicellular developmental diversity of protists
www.dudinlab.com
#Ichthyosporea, #Multicellularity, #Evolution, #Embryo, #Development, #Protist, #UExM, #Cytoskeleton, #Expansion #Microscopy
I love science! I study toxin interactions in microbes. Dad of two @wellcometrust Sir Henry Wellcome Fellow | All views my own | he/him | 🏳️🌈
Mom of two ❤️
Early-career researcher in AMR, Mobile Genetic Elements, and One Health & Part-time lecturer.
In love with the intelligence of microbes, their evolutionary designs, and how they outthink every threat.
JUST- Jordan 🇯🇴
Stanford Biophysics PhD Student with Dmitri Petrov and Ben Good. evolution, ecology, genomics, microbiomes, biophysics :) she/her
https://sophiejwalton.github.io
Post-doc at D-HEST, ETH Zurich. PhD in microbiology at Biozentrum, Basel. Phagehunter. Hiker. Scout. Music and food enthusiast. Per aspera ad astra
https://orcid.org/0000-0002-0751-9572
Microbiologist |microbial ecology | microbiome | phage-bacteria interactions
phages rule the world👀
Postdoc at Evolutionary Biology lab, Gulbenkian Institute for Molecular Medicine - Oeiras
MRC Career Development Award fellow at the University of Cambridge | Group Leader of the Microbiome Function and Diversity lab (https://microfundiv-lab.github.io) | Bioinformatics, Machine Learning, Metagenomics, Functional modelling, Human microbiome