Finally, I’m still on lookout for my next opportunity and would love to connect with anyone in industry/biotech space! Feel free to reach out here or on the floor!
#AAIC #AAIC25
27.07.2025 13:42 — 👍 3 🔁 1 💬 0 📌 0
Come check out our whole session (starts at 4:15pm) with excellent talks from collaborators Evan Macosko, Henrik Zetterberg, Ville Leinonen, & Tarja Malm.
𝙎𝙩𝙪𝙙𝙮𝙞𝙣𝙜 𝙩𝙞𝙨𝙨𝙪𝙚 𝙨𝙖𝙢𝙥𝙡𝙚𝙨 𝙛𝙧𝙤𝙢 𝙧𝙖𝙧𝙚 𝙘𝙤𝙝𝙤𝙧𝙩𝙨 𝙩𝙤 𝙡𝙚𝙖𝙧𝙣 𝙖𝙗𝙤𝙪𝙩 𝙚𝙖𝙧𝙡𝙮 𝙨𝙩𝙖𝙜𝙚 𝘼𝘿 𝙥𝙖𝙩𝙝𝙤𝙥𝙝𝙮𝙨𝙞𝙤𝙡𝙤𝙜𝙮.
27.07.2025 13:42 — 👍 2 🔁 0 💬 1 📌 0
Excited for start of AAIC!
I’ll be giving talk Tuesday afternoon on unique population of AD-relevant macrophages we have identified in CSF via scRNA-seq:
𝗦𝗶𝗻𝗴𝗹𝗲 𝗖𝗲𝗹𝗹 𝗧𝗿𝗮𝗻𝘀𝗰𝗿𝗶𝗽𝘁𝗼𝗺𝗶𝗰𝘀 𝗼𝗳 𝗣𝗮𝗶𝗿𝗲𝗱 𝗖𝗦𝗙 𝗮𝗻𝗱 𝗕𝗹𝗼𝗼𝗱 𝗜𝗺𝗺𝘂𝗻𝗲 𝗣𝗼𝗽𝘂𝗹𝗮𝘁𝗶𝗼𝗻𝘀 𝗥𝗲𝘃𝗲𝗮𝗹𝘀 𝗨𝗻𝗶𝗾𝘂𝗲 𝗠𝗮𝗰𝗿𝗼𝗽𝗵𝗮𝗴𝗲 𝗣𝗼𝗽𝘂𝗹𝗮𝘁𝗶𝗼𝗻 𝘄𝗶𝘁𝗵 𝗥𝗲𝗹𝗲𝘃𝗮𝗻𝗰𝗲 𝘁𝗼 𝗔𝗹𝘇𝗵𝗲𝗶𝗺𝗲𝗿'𝘀 𝗗𝗶𝘀𝗲𝗮𝘀𝗲
27.07.2025 13:42 — 👍 4 🔁 1 💬 1 📌 0
scCustomize 3.0.1 now on CRAN. Hotfix release with couple of bug fixes.
Enjoy!
19.12.2024 12:43 — 👍 5 🔁 1 💬 0 📌 0

a cartoon character is holding a can of beans with the words how do i use it below him
ALT: a cartoon character is holding a can of beans with the words how do i use it below him
Implementation and validation of single-cell genomics experiments in neuroscience
www.nature.com/articles/s41...
Validation is key! This review dives into best practices for validating ssc/snRNAseq experiments. Learn about the challenges and solutions for ensuring robust and reliable findings.
11.12.2024 17:04 — 👍 7 🔁 2 💬 1 📌 0

3 reviews on planning, execution, & validation of transcriptomic experiments in neuroscience published in @natureportfolio.bsky.social #NatureNeuroscience.
A MUST READ if you're doing transcriptomics.
3 reviews, an amazing editorial, a beautiful cover – this is the issue of the year!
(mini thread)
11.12.2024 17:04 — 👍 118 🔁 44 💬 2 📌 3
Finally, as always, I want to thank everyone who uses scCustomize. I could never have imagined it would be adopted and used by some many people and it’s really fantastic every time I read paper or see plot that was made using scCustomize.
Happy single cell analysis!!! 💻📈🧬
20/20
06.12.2024 16:36 — 👍 1 🔁 0 💬 0 📌 0
As always there are a ton of bug fixes in this update here too. Thank you to everyone who has report issues on GitHub!
There’s more in this update too besides these highlights so check out changelog for full details!!
19/n
06.12.2024 16:36 — 👍 1 🔁 0 💬 1 📌 0
A number of functions have updated parameters, including `Clustered_DotPlot` which has 8!! new parameters to further customize the output plot and plot legends.
See changelog for full details on new/updated parameters in this update.
18/n
06.12.2024 16:36 — 👍 0 🔁 0 💬 1 📌 0
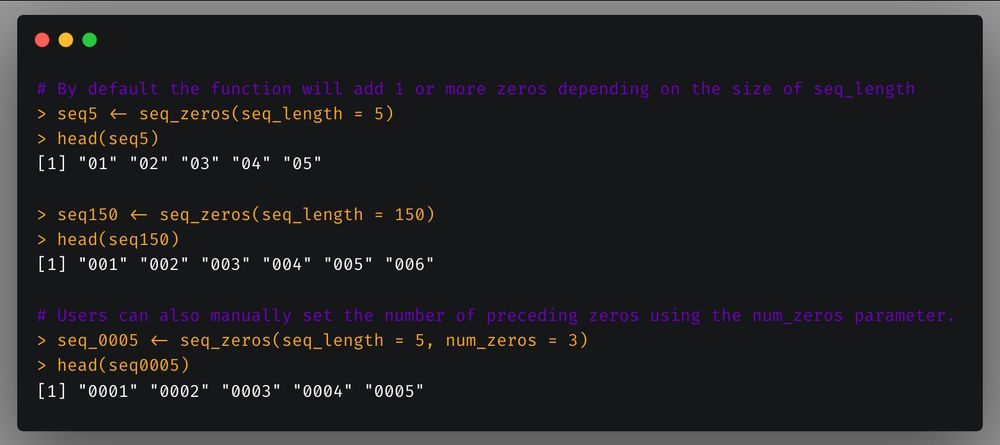
scCustomize now includes `seq_zeros` function to easily create sequences with preceding zeros. You can either specify the number of preceding zeros desired or the function will set automatically based on sequence length.
17/n
06.12.2024 16:36 — 👍 0 🔁 0 💬 1 📌 0
The base R `seq` family of functions has a ton of uses. However, due to the way numbers are ordered in R (and other software) it can sometimes be helpful to have preceding zeros in your number sequences in order to keep things in numerical order (e.g., 01, 02, 03, instead of 1, 2, 3).
16/n
06.12.2024 16:36 — 👍 0 🔁 0 💬 1 📌 0
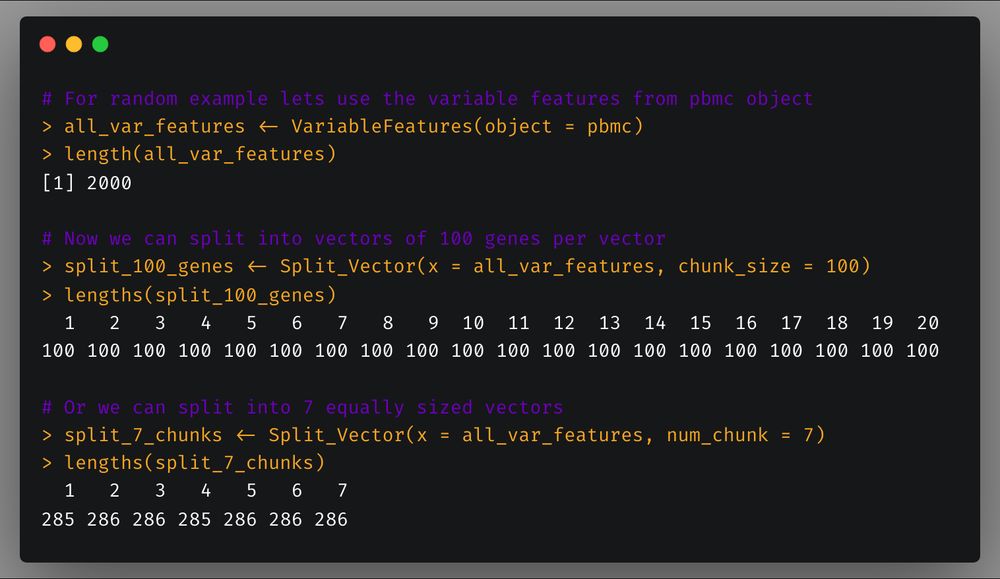
Now here’s two functions that aren’t limited to scRNA-seq analysis but can generally be helpful in your work in R.
First, `Split_Vector` which allows you to split a vector in a specified number of chunks or chunks of specific length.
15/n
06.12.2024 16:36 — 👍 0 🔁 0 💬 1 📌 0
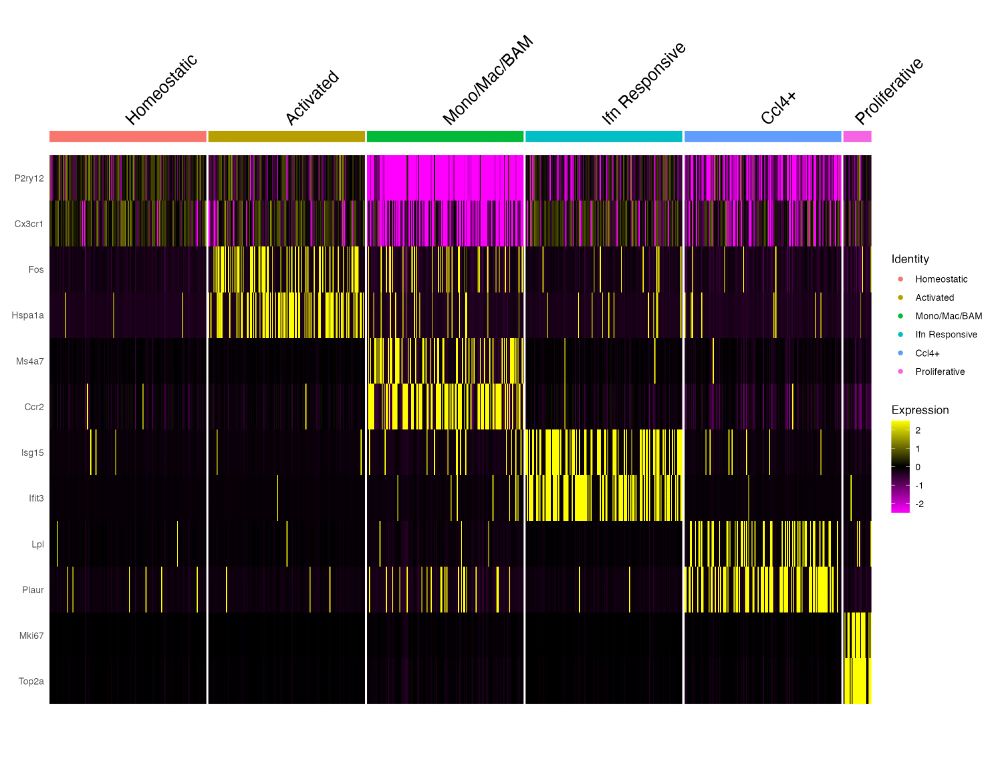
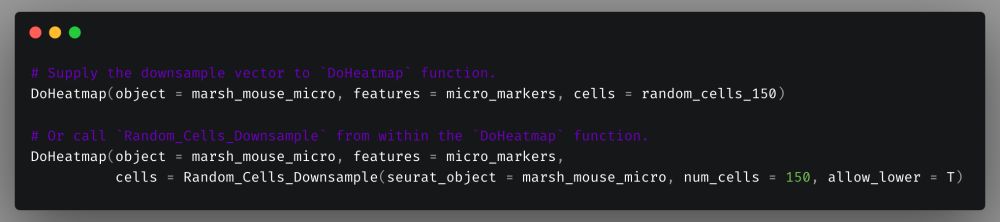
Now we supply that to `cells` parameter of `DoHeatmap` and the result now looks like this. We can now clearly visualize the differences in expression between the identities.
To make it even easier you can also just call `Random_Cells_Downsample` from within the `DoHeatmap` call.
14/n
06.12.2024 16:36 — 👍 0 🔁 0 💬 1 📌 0
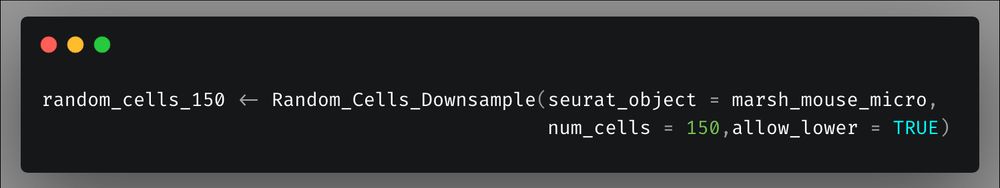
However, by using `Random_Cells_Downsample` we can create a plot with equal number of cells per identity for proper visualization. Here I create a down sampled cell vector of 150 cells per group (or the max number of cells for groups smaller than 150 cells)
13/n
06.12.2024 16:36 — 👍 0 🔁 0 💬 1 📌 0
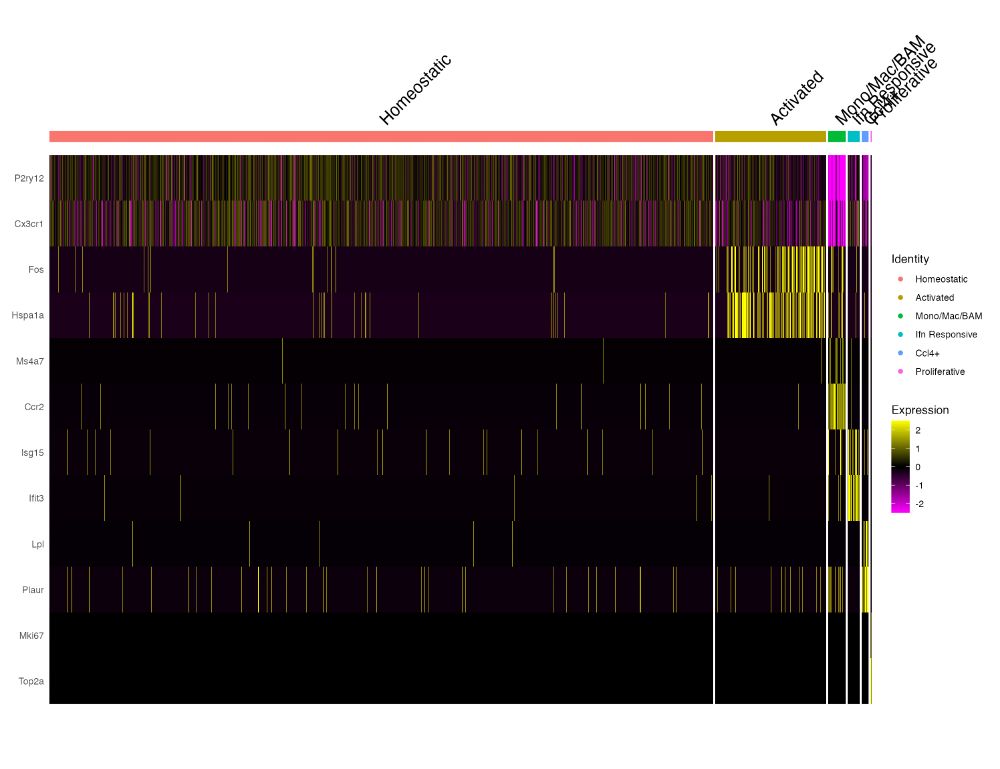
The default cell level heatmap scales the size of each identity based on the number of cells. This can lead to heatmaps where visualizing the expression in small clusters can be nearly impossible.
12/n
06.12.2024 16:36 — 👍 0 🔁 0 💬 1 📌 0
This update also contains new feature called `Random_Cells_Downsample` to return randomly downsampled set of cells from object with equal numbers of cells per identity.
There are many scenarios where this can be helpful but one that I use a lot is when plotting cell-level heatmaps.
11/n
06.12.2024 16:36 — 👍 0 🔁 0 💬 1 📌 0
Just like human function, `Updated_MGI_Symbols` requires one time internet connection to download the MGI file but then stores it in local cache so not internet is required for subsequent use.
10/n
06.12.2024 16:36 — 👍 0 🔁 0 💬 1 📌 0
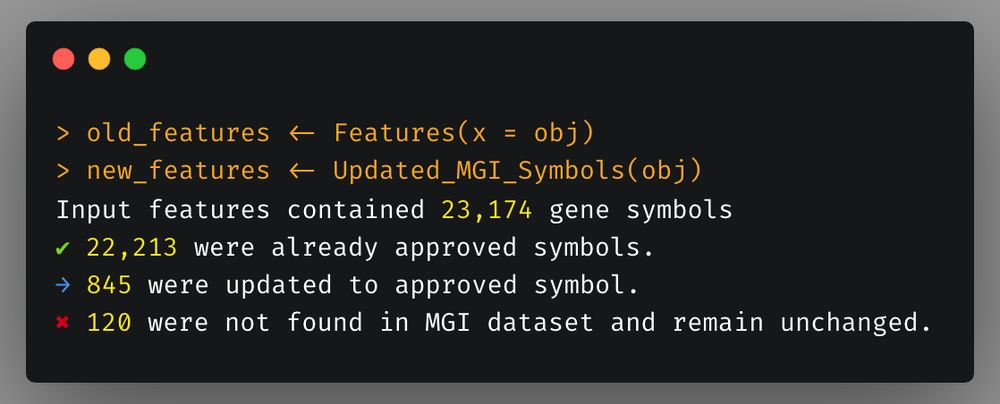
The last scCustomize update brought the advance of `Updated_HGNC_Symbols` for updating human gene symbols. This update brings the equivalent function of mice using MGI database: `Updated_MGI_Symbols`.
9/n
06.12.2024 16:36 — 👍 0 🔁 0 💬 1 📌 0
Plotting #5: Spatial Plotting Functions
scCustomize
scCustomize also makes it’s first venture in spatial plotting with addition of `SpatialDimPlot_scCustom` which is sibling to `DimPlot_scCustom` and contains similar updates to color scheme and other defaults.
See new spatial vignette for details:
samuel-marsh.github.io/scCustomize/...
8/n
06.12.2024 16:36 — 👍 0 🔁 0 💬 1 📌 0
One new plotting function “Factor_Cor_Plot” has been added which will plot correlation between feature loadings of liger iNMF factors (works with both Seurat and Liger objects).
LIGER vignette is in progress and will I get that finished as soon as I can!
7/n
06.12.2024 16:36 — 👍 0 🔁 0 💬 1 📌 0
Old liger functions have been updated to enable working with either old or new style liger objects.
New liger interaction functions have been added. “Subset_LIGER”, “Cells_by_Identities_LIGER”.
6/n
06.12.2024 16:36 — 👍 0 🔁 0 💬 1 📌 0
scCustomize v3.0.0 contains massive updates for interaction with new style liger objects (liger v2.0.0+). Changelog contains the full function update list but some highlights are:
Extending Seurat generic functions to work with liger (Cells, Features, Idents, WhichCells, etc)
5/n
06.12.2024 16:36 — 👍 0 🔁 0 💬 1 📌 0
Object QC Functions
scCustomize
In addition to new species and metrics `Add_Cell_QC` is now S3 generic function and works with either Seurat or Liger objects.
I have also added new vignette specifically dedicated to this and other QC functions in scCustomize.
samuel-marsh.github.io/scCustomize/...
4/n
06.12.2024 16:36 — 👍 0 🔁 0 💬 1 📌 0
`Add_Cell_QC` also can work with ensembl IDs instead of feature names and has all ensembl IDs for default species stored within the package, no download required!
3/n
06.12.2024 16:36 — 👍 0 🔁 0 💬 1 📌 0
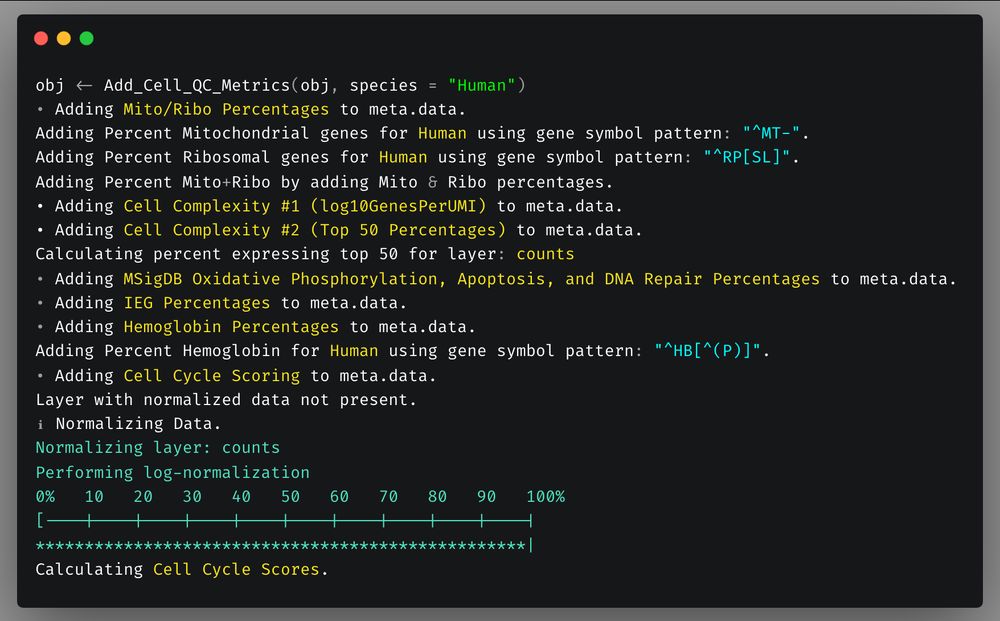
First, QC functions.
`Add_Cell_QC_Metrics` now includes addition of % of hemoglobin counts as metric & chicken was added to default species.
Default species are now human, mouse, rat, drosophila, zebrafish, macaque, marmoset, & chicken.
One line will add all QC metrics to your object.
2/n
06.12.2024 16:36 — 👍 0 🔁 0 💬 1 📌 0
So excited to share our work developing a new class of modular synthetic GPCRs, out today @Nature! Check it out nature.com/articles/s41...
05.12.2024 08:10 — 👍 33 🔁 13 💬 3 📌 1
Could one envision a synthetic receptor technology that is fully programmable, able to detect diverse extracellular antigens – both soluble and cell-attached – and convert that recognition into a wide range of intracellular responses, from gene expression and real-time fluorescence to modulation..
04.12.2024 16:05 — 👍 434 🔁 138 💬 33 📌 17
Nature Neuroscience - Focus on single-cell genomics in neuroscience
Single-cell and single-nucleus genomics pave the way for a comprehensive understanding of the nervous system and its diverse cell populations. Realizing this...
Spotlight on Single-Cell Genomics!
I, Omer Ali Bayraktar, Naomi Habib, Genevieve Konopka, @liddelowsa.bsky.social, and Tomasz Nowakowski collaborating with top neuroscientists to explore optimizing these technologies in the latest Nature Neuroscience issue.
🔗 www.nature.com/neuro/volume...
03.12.2024 21:04 — 👍 58 🔁 21 💬 2 📌 1
Nature Neuroscience - Focus on single-cell genomics in neuroscience
Single-cell and single-nucleus genomics pave the way for a comprehensive understanding of the nervous system and its diverse cell populations. Realizing this...
The December issue of Nature Neuroscience is now live, featuring our Focus on Single-Cell Genomics! We hope that these consensus reviews from experts in the field will serve as a useful guide for neuroscientists regarding best practices for these approaches.
www.nature.com/neuro/volume...
03.12.2024 19:49 — 👍 82 🔁 21 💬 1 📌 0
Neuroscientist at College de France in Paris🧠🔬👩🔬
Interested in neuron-glia interactions during behavior
Exploring how the environment, brain & body interact to shape human health 🔬 | Faculty at Stanford & Core Investigator at @arcinstitute.org
http://www.thaisslab.com
Postbac fellow | neuroglia 🌟 | future PhD student 🤞🏻🧠 | uva alum
Postdoc in the Yang Lab at the Gladstone Institutes
Engineering cells to understand their decisions
ESPOD Fellow @ebi.embl.org & @sangerinstitute.bsky.social
(Saez-Rodriguez & Parts)
lingering scientist @crick.ac.uk (Briscoe)
Postdoctoral Researcher | Neurodegeneration, Lysosomes & FTD | Rosa Rademakers Lab, VIB-UAntwerp Center for Molecular Neurology
Group of passionate scientists unraveling the mysteries of neurodegeneration at Weill Cornell Medicine 🧠 | Tackling Alzheimer’s, FTD, PD & related diseases 🧬 | Focused on tau, innate immunity, & targeted therapeutics using iPSC & transcriptomics🔬
Research Associate, University of Cambridge | Leading Edge fellow | first-gen academic | molecular biologist studying effects of stress across the lifespan 🧠🩸🧬
The Alzheimer’s Association is the leading voluntary health organization in Alzheimer's care, support and research. #ENDALZ
alz.org
🇦🇫 PhD scientist @ucdavis SOM | Bioinformatics | Neuroscience | Neurodevelopment | Epigenetics | -Omics
Scientist-in-training, Ph.D. student in Kashkar lab 👩🏻🔬 at the University Hospital Cologne @CECAD
Passionate about cell death and inflammation in eye and brain🔬🧬🧠🎼
Postdoc in @liganlab.bluesky.social at Weill Cornell Medicine interested in resilience and microglia in neurodegeneration • Vanderbilt Neuro PhD ‘22 • Kenyon College ‘17 • 1st gen Egyptian-American
UCSD Neuroscience PhD Student | Salk Institute Allen Lab | UC Davis Olson Lab Alum | she/her
Incoming assistant professor @UAMS
Neural stem cells & the choroid plexus in neuroinflammation using patient-based 2D and 3D models
Senior Research Associate, Dept of Clinical Neurosciences, University of Cambridge
translational cell biologist| senior postdoctoral scientist | proteostasis & neuroinflammation | vrije universiteit brussel (VUB)
PhD candidate | Vollum Institute | Bret Pearson Lab | Neural regeneration
Single-Cell & Spatial Transcriptomics | Group Leader at GliaOmics Lab | Head of GeneCore Facility | Co-founder at Carta Genum | Glial Cells in Health & Disease
Child Lung specialist, Queensland Children’s Hospital. Conjoint Prof, Uni Queensland & Director Children's Health & Environment Program (CHEP). International Conference Committee Chair-elect Amer Thoracic Soc. My views on lung health #MedSky #PedsPulmSky
Neuroimmunology Lab (NIM) at VIB-UAntwerp CMN
We study the interactions between the immune and nervous systems in neurodegenerative and neurodevelopmental disorders.
https://pasciutolab.sites.vib.be/en
Postdoc, works on C9ALS/FTD @MancusoRenzo @VIB_CMN










