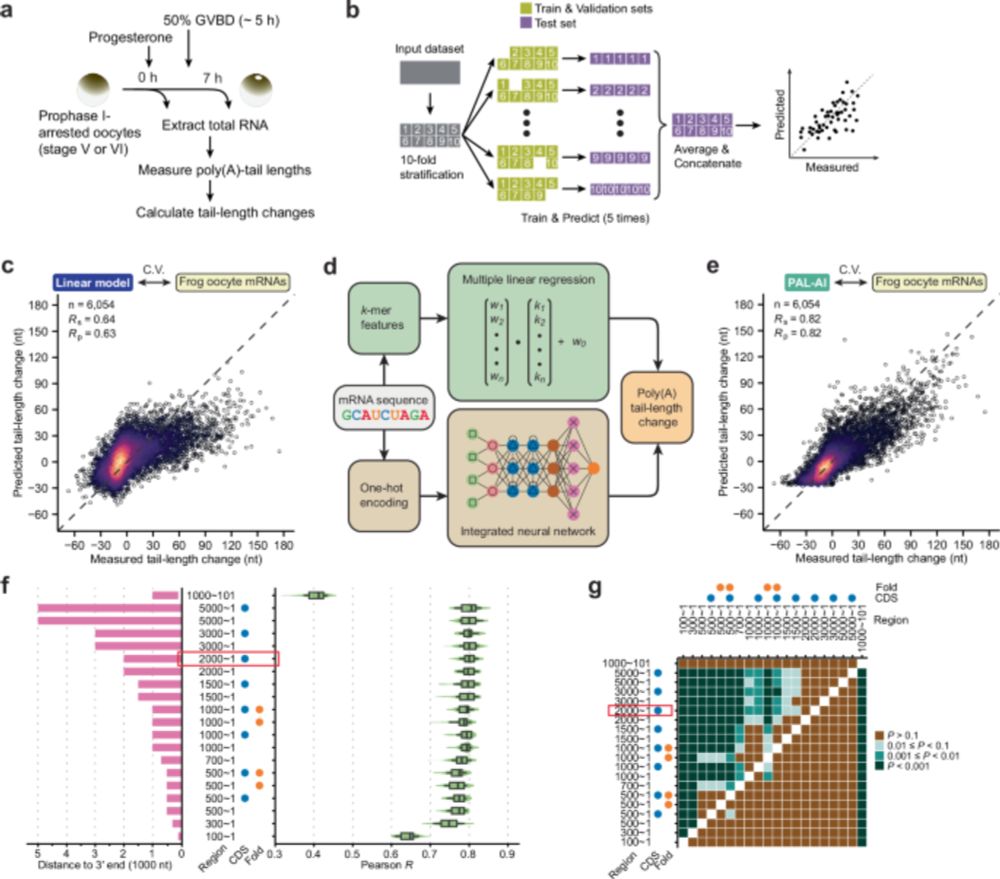
🧪🧬New preprint We present cryo-EM structures of reconstituted CTCF–nucleosome complexes, showing CTCF dimerization drives nucleosome oligomerization into defined higher-order assemblies. Disrupting CTCF–CTCF interfaces in mESCs reduces looping and impairs differentiation. tinyurl.com/CTCF-nucleos...
09.02.2026 12:54 — 👍 102 🔁 48 💬 4 📌 2
We’re excited to share our latest preprint on the mechanism of excised linear intron stabilization in yeast! This work was led by PhD student @glennli.bsky.social and was a wonderful collaboration with @maxewilkinson.bsky.social. Link: www.biorxiv.org/content/10.6... (1/4)
23.01.2026 16:14 — 👍 58 🔁 25 💬 1 📌 2

Check our latest collaboration with the Kleaveland Lab (kleavelandlab.org), led by Joanna Stefano and Lara Elcavage: academic.oup.com/nar/article/... (1/2)
14.01.2026 21:11 — 👍 14 🔁 4 💬 1 📌 1

Stepwise DNA unwinding gates TnpB genome-editing activity
TnpB is a compact RNA-guided endonuclease and evolutionary ancestor of CRISPR-Cas12 that offers a promising platform for genome engineering. However, the genome-editing activity of TnpBs remains limit...
New preprint 👉Doudna x Bryant x Jacobsen x Savage collaboration!
Work led by @zehanzhou.bsky.social, I. Saffarian-Deemyad, @honglue.bsky.social, T. Weiss
We dissect how stepwise DNA unwinding gates TnpB genome editing, revealing how unwound DNA states enhance cleavage
www.biorxiv.org/content/10.6...
10.01.2026 17:16 — 👍 25 🔁 11 💬 0 📌 0
When RNA Degradation 🤝 meets 🤝 Protein Degradation! tinyurl.com/E3TDMD In a collaboration of @bartellab.bsky.social and Schulman lab, we show that, in target-directed microRNA degradation (TDMD), 2-RNA-factors recruit an E3 ligase and induce the degradation of not only a protein but also RNA (1/5).
06.01.2026 08:04 — 👍 115 🔁 50 💬 1 📌 4
an interlocked dimer, or that RNA recognition would be part of the mechanism, or that ZSWIM8 had found its way into AGO's PAZ pocket. Very glad for this to be shared with the world today. Congrats to everyone on the team @jakobfarnung.bsky.social @elenaslo.bsky.social et al! [2/2]
06.01.2026 18:33 — 👍 4 🔁 0 💬 0 📌 0
It's finally the day we unveil this beautiful mechanism + structure of TDMD! It's been a most riveting, collaborative journey with the team. I remember every exciting moment that we revealed a new small piece of the puzzle over the past years, e.g. when we found out that ZSWIM8 would be [1/2]
06.01.2026 18:33 — 👍 9 🔁 0 💬 1 📌 0
✨New preprint!
🧵1/4 Excited to share our work on AI-guided design of minimal RNA-guided nucleases. Amazing work by @petrskopintsev.bsky.social @isabelesain.bsky.social @evandeturk.bsky.social et al!
Multi-lab collaboration @banfieldlab.bsky.social @jhdcate.bsky.social @jacobsenucla.bsky.social🧬
🔗👇
09.12.2025 07:52 — 👍 98 🔁 47 💬 1 📌 8
My first first-author paper is out!🎉
Here we propose a model where a silencing complex, PIWI*, assembles on target RNAs to recruit effectors and shut down transposon activity.
Huge thanks to the Brennecke and Plaschka labs, especially Julius and Clemens, and all co-authors!
17.09.2025 13:00 — 👍 48 🔁 20 💬 3 📌 1
Congrats!!
17.09.2025 14:01 — 👍 1 🔁 0 💬 0 📌 0
Congrats Jimmy!
11.09.2025 00:18 — 👍 1 🔁 0 💬 0 📌 0
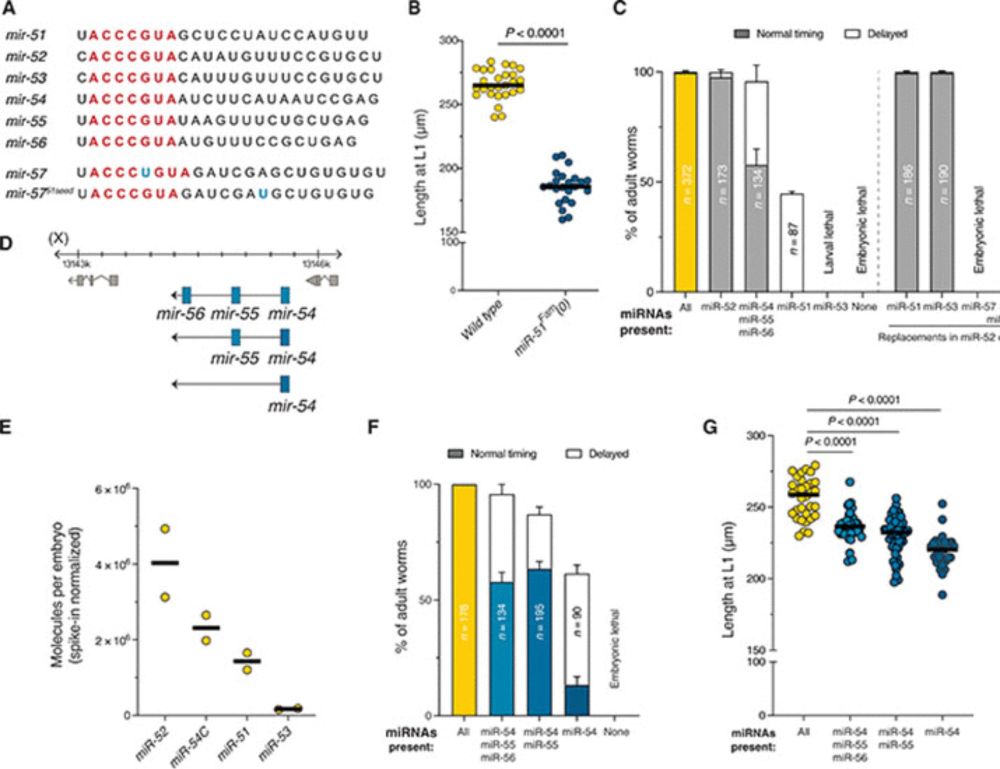
An ancient and essential miRNA family controls cellular interaction pathways in C. elegans
A microRNA that arose at the origin of eumetazoans regulates cell adhesion and signaling in C. elegans through conserved targets.
Our work on the function of miR-51/miR-100 is out! miR-100 is widely conserved across eumetazoans but its function has been mysterious. Emilio Santillán found in worms it regulates signaling and extracellular matrix genes, some of which seem to be conserved targets! www.science.org/doi/10.1126/...
03.09.2025 18:16 — 👍 93 🔁 35 💬 5 📌 7

Argonautes are coming :-) ... soon at @imgprague.bsky.social ... finishing preparations. Still missing a few things like drums, guitar, badges, beer ... but it's under control. I think. I hope. :-))
#argonautes2025
21.08.2025 12:48 — 👍 37 🔁 9 💬 1 📌 2
Why would anyone want to be a scientist?
It is difficult to fathom why anyone intelligent enough to be a scientist would actually choose to be one. Doing good science requires the utmost exertion of body, mind and spirit, yet is consistently...
"why [would] anyone intelligent enough to be a scientist choose to be one [given] the unfavorable risk-to-reward ratio "?
One of the most intelligent people you could meet offers some answers: having ideas, watching them develop, and sharing them journals.biologists.com/jcs/article/...
15.08.2025 16:16 — 👍 117 🔁 40 💬 5 📌 8
AlphaFold is amazing but gives you static structures 🧊
In a fantastic teamwork, @mcagiada.bsky.social and @emilthomasen.bsky.social developed AF2χ to generate conformational ensembles representing side-chain dynamics using AF2 💃
Code: github.com/KULL-Centre/...
Colab: github.com/matteo-cagia...
17.04.2025 19:10 — 👍 205 🔁 63 💬 3 📌 4
Don’t miss this Q&A with Dr. Michelle Frank (@michelle-frank.bsky.social), an awesome postdoc in our lab!
05.04.2025 19:51 — 👍 18 🔁 2 💬 0 📌 0

A miniature CRISPR-Cas10 enzyme confers immunity by an inverse signaling pathway
Microbial and viral co-evolution has created immunity mechanisms involving oligonucleotide signaling that share mechanistic features with human anti-viral systems. In these pathways, including CBASS a...
Preprint alert! ✨ In this project that I co-led with @benadler.bsky.social, we show that a miniature CRISPR-Cas10-like enzyme, mCpol, uses a novel inverse signaling mechanism to prevent the spread of viruses that attempt immune evasion by depleting host cyclic nucleotides.
Check it out:
31.03.2025 16:27 — 👍 63 🔁 30 💬 4 📌 3
Congrats Jimmy and team! Excited to see this work out
28.03.2025 12:06 — 👍 1 🔁 0 💬 1 📌 0
Congrats Leo and all!
26.03.2025 13:51 — 👍 3 🔁 0 💬 0 📌 0
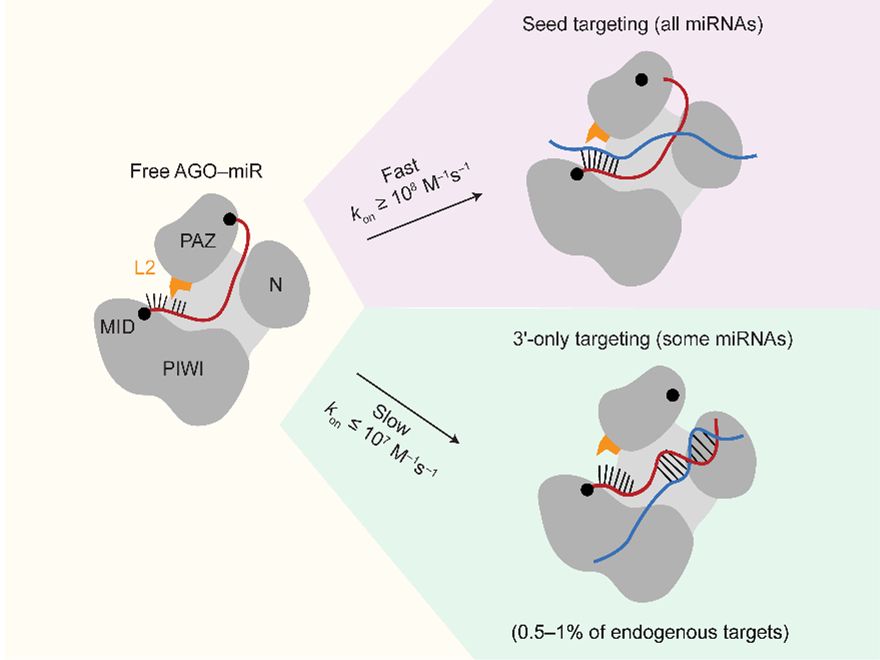
Check out the latest study from our lab, led by @mhall98.bsky.social :
www.biorxiv.org/content/10.1... (1/2)
15.03.2025 00:36 — 👍 48 🔁 17 💬 1 📌 1
Check out the latest work led by @mhall98.bsky.social in the Bartel lab! Matt solved a long-standing mystery about miRNA targeting, and drove the story to impressive mechanistic detail. More information below:
13.03.2025 15:44 — 👍 3 🔁 0 💬 0 📌 0

ARGONAUTES 2025
The grassroot meeting of Argonaute afficionados is looking for afficionados! Full coverage of Argonautes from prokaryotes to #RNAi, #piRNA and #microRNA Join us in Prague at @imgprague.bsky.social during 27-30/8, 2025. Registration, program and more info here:
argonautes.img.cas.cz
12.03.2025 06:56 — 👍 18 🔁 14 💬 1 📌 6

Today we're thrilled to announce our 2024 #HannaGrayFellows! Please join us in welcoming and celebrating these outstanding early career scientists!
08.01.2025 14:24 — 👍 118 🔁 45 💬 1 📌 37
Our research group is interested in the molecular and cellular origins and evolution of vertebrate organs. Tweets by Henrik, usually in the name of our group.
home.kaessmannlab.org
The longest running international CRISPR conference, established in 2008. CRISPR 2026 will be held from June 10 - 12 in New Haven, Connecticut, USA
PhD Candidate at Bartel Lab (Whitehead Institute and MIT).
Scientist interested in genes in living cells. Associate Professor at Colorado State University.
studying genetic and environmental contributions to cell fitness in cancer and immunity | biology-engineering-biotechnology | #HPLM | current: investigator @Morgridge_Inst, assistant prof @UW-Madison
Compartmentalizing your metabolism, and y’know, CRISPR stuff. Account managed by grad students and postdocs 🤙
@ucberkeleyofficial.bsky.social @innovativegenomics.bsky.social @hhmi.bsky.social
savagelab.org
Interest in all things transposon + RNA + cancer biology!
PhD candidate @ UC Berkeley
Characterization & delivery of CRISPR proteins
Assistant Professor at RNA Therapeutics Institute, UMass Chan Medical School
Molecular machines, Proteostasis, Ubiquitin Signaling, Cell Autonomous Defense, Protein Disorders and Team Nematode
@IMP Vienna
www.imp.ac.at/groups/tim-clausen/
Studying genetic interactions in cancer cells using CRISPR-based functional genomics approaches at NCI/NIH. Views are my own.
Asst. Prof. Uni Groningen 🇳🇱
Comp & Exp Biochemist, Protein Engineer, 'Would-be designer' (F. Arnold) | SynBio | HT Screens & Selections | Nucleic Acid Enzymes | Biocatalysis | Rstats & Datavis
https://www.fuerstlab.com
https://orcid.org/0000-0001-7720-9
MPhil student @ Cambridge history of science | via @doudna-lab.bsky.social, Princeton/Bassler Lab | Demystifying biotech and biosecurity at scifinow.substack.com/ 🧬🧪☣️
PostDoc @Doudna_lab 🧬| PhD @Cambridge_Uni | @Harvard @imperialcollege - I like epigenetics and editing genes. Life begins at the end of your comfort zone✨✈️🌍⛵️🎿
News from the Rubin Lab 🧬🧫 We conduct DNA-level editing of microbial communities for understanding and control. Managed by lab members.
@innovativegenomics.bsky.social @ucberkeleyofficial.bsky.social
🔗 therubinlab.org
Principal Investigator at @aithyra.bsky.social | Structural bioinformatics, virology, and innate immunity | jasonnomburg.com
Ribosome is life. Posts by grads/postdocs, approved (sometimes) by Rachel Green.
@Johns Hopkins Medical Institute
https://greenlabjhmi.org
signaling systems and protein circuitry. reimagining what cells can be. fun posts only. Assistant Professor:
@uwbiochem | Postdoc: @stanford @prakashlab | Ph.D.:
@ucsf Wendell Lim @CDI_UCSF
Research and development company leveraging the biology of diverse organisms. Read about our science: http://research.arcadiascience.com