
Stoked to finally have a preprint out for Phold, our tool that uses protein structural information to enhance phage genome annotation #phagesky 1/n
www.biorxiv.org/content/10.1...
@hannelorelongin.bsky.social
PhD student @ Computational Systems Biology & Lab of Gene Technology. Investigating PTMs in viruses and bacteria with bioinformatics 💻🦠

Stoked to finally have a preprint out for Phold, our tool that uses protein structural information to enhance phage genome annotation #phagesky 1/n
www.biorxiv.org/content/10.1...

🚨 New preprint 🚨
My phage annotation tool, Phynteny, finally has a preprint and a brand new version powered by a cool AI transformer architecture and protein language models! #phagesky
www.biorxiv.org/content/10.1...
Curious about our findings and where we're headed? Happen to be at #ISMB2025 ? Come check out
my poster (A-229, function COSI) today!

📣 New preprint alert!
Happy to share the first results from our effort to reannotate Pseudomonas #phage proteins of unknown function, using structural bioinformatics.
📄 doi.org/10.1101/2025...
We've folded a lot of proteins! You can see some of them here @linsalrob.bsky.social @gbouras13.bsky.social
07.07.2025 08:49 — 👍 13 🔁 5 💬 0 📌 0Hip hip hooray: our #PTM conservation tool FLAMS turns 1 🎂
To celebrate, we have added handy new features to FLAMS web, incl. 3D visualization of hit proteins & PTM sites with known active/binding sites.
Come check it out @ tinyurl.com/flamsweb & let us know what you think!
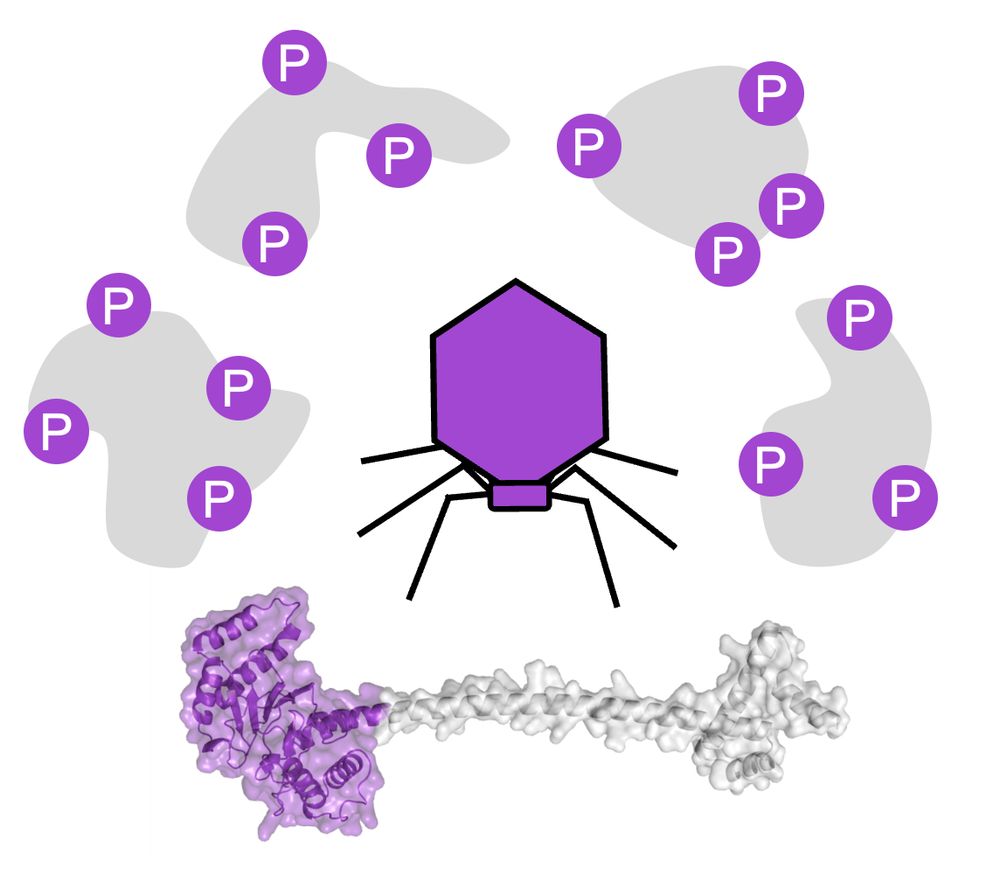
1/11 - With Karin Mitosch and Clément Potel + colleagues in Savitski lab and @typaslab.bsky.social @embl.org, I’m very happy to share our discovery of a hyper-promiscuous phage protein kinase (T7K) which broadly deactivates bacterial defenses! 🦠
www.biorxiv.org/content/10.1...