Happy to share part of my postdoctoral work at the @lucas.farnunglab.com lab. Great collaboration with @voslab.org and @andersshansen.bsky.social. “Structural basis for CTCF-mediated chromatin organization” www.biorxiv.org/content/10.6...
09.02.2026 15:22 — 👍 30 🔁 10 💬 0 📌 0
🧪🧬New preprint We present cryo-EM structures of reconstituted CTCF–nucleosome complexes, showing CTCF dimerization drives nucleosome oligomerization into defined higher-order assemblies. Disrupting CTCF–CTCF interfaces in mESCs reduces looping and impairs differentiation. tinyurl.com/CTCF-nucleos...
09.02.2026 12:54 — 👍 115 🔁 50 💬 4 📌 3
🧬📄 Preprint update (v2): We added new biochemical experiments that clarify how IWS1 associates with the transcription elongation complex and further define competition between IWS1 and RECQL5. 🧪⚙️
www.biorxiv.org/content/10.1...
Structural model is already available in the PDB (9MLC).
02.02.2026 16:33 — 👍 47 🔁 15 💬 1 📌 0
The two that first showed us how FACT engages nucleosomes now have their own lab and show us how the remodeler ATRX engages nucleosomes! Incredible resolution with a lot of biochemistry, congrats @codyz.bsky.social @yangliu9.bsky.social and team!
28.01.2026 11:18 — 👍 4 🔁 1 💬 1 📌 0
My first research paper as an independent PI is now online. Thank you to the authors for contributions and to the reviewers for thoughtful and constructive comments. Grateful to the editor for the opportunity. onlinelibrary.wiley.com/doi/10.1002/...
17.01.2026 16:29 — 👍 3 🔁 1 💬 0 📌 1

What an exciting meeting at this month’s Gene Expression and RNA Series (GEARS) in Boston! Thank you to everyone who joined us for an introduction to the HCR™ platform.
Special shoutout to @xiaoliwu.bsky.social, jonmarkert.bsky.social, and @ara-latifkar.bsky.social for helping coordinate the event!
24.11.2025 20:10 — 👍 1 🔁 1 💬 0 📌 1

Please join us on November 20th! We are very grateful for our first sponsorship - Molecular Instrument!
18.11.2025 19:39 — 👍 1 🔁 3 💬 0 📌 0
Congrats CJ and team!!
30.10.2025 22:08 — 👍 1 🔁 0 💬 1 📌 0
🧬 Transcription elongation by RNA polymerase II relies on a web of elongation factors. Our new work shows how IWS1 acts as a modular scaffold to stabilize & stimulate elongation. Fantastic work by Della Syau! www.biorxiv.org/content/10.1...
29.08.2025 10:14 — 👍 57 🔁 23 💬 1 📌 1
YouTube video by Fragile Nucleosome
Jon Markert
Check out @jonmarkert.bsky.social's recent talk for the @fnucleosome.bsky.social series, covering his recent breakthrough paper (www.science.org/stoken/autho...) on co-transcriptional histone mark deposition of H3K36me3: www.youtube.com/watch?v=ByJr...
30.05.2025 11:21 — 👍 19 🔁 4 💬 0 📌 0

🧬🎉Thrilled to share our new chromatin remodeling study! We reveal three states of human CHD1 and identify a novel "anchor element" that interacts with the acidic patch—conserved among remodelers. Our structures clarify mechanisms of remodeler recruitment! Link: authors.elsevier.com/a/1l2ik3vVUP...
06.05.2025 15:08 — 👍 103 🔁 24 💬 4 📌 3

Excited to see everyone tonight at Whitehead Institute for the 2nd GEARS seminar, featuring three talks from Harleen Saini, Vikram Agarwal, and Ava Carter!
01.05.2025 20:09 — 👍 5 🔁 1 💬 0 📌 1
I’m excited to share my first paper in the Farnung lab, in which we report four cryo-EM structures of the human DNA replication machinery engaging with and progressing into a nucleosome.
06.04.2025 10:43 — 👍 47 🔁 13 💬 4 📌 0
Our work on DNA replication is out! Led by @felixsteinruecke.bsky.social, we monitor the replisome progressing into nucleosomes!
06.04.2025 10:51 — 👍 13 🔁 1 💬 0 📌 0
Take a look at our new collaboration project with the Phil Cole lab, we demonstrate how SIRT6 can recognize and remove a variety of different Histone acylations!
28.03.2025 16:16 — 👍 10 🔁 4 💬 0 📌 0
Are you interested in hearing this SETD2 story in more detail? I’ll be presenting it at @fnucleosome.bsky.social this week! Please see the poster for more details
10.03.2025 14:20 — 👍 8 🔁 2 💬 0 📌 0

🥁We are just 3 days away from our next #FragileNucleosome seminar! We are delighted to host, 2 amazing ECR researchers @rachelhoffman.bsky.social & @jonmarkert.bsky.social this Wed!
PS1: Don't forget that US has switched to PDT
PS2: The recurring registration link:
us06web.zoom.us/webinar/regi...
10.03.2025 13:28 — 👍 12 🔁 11 💬 0 📌 4
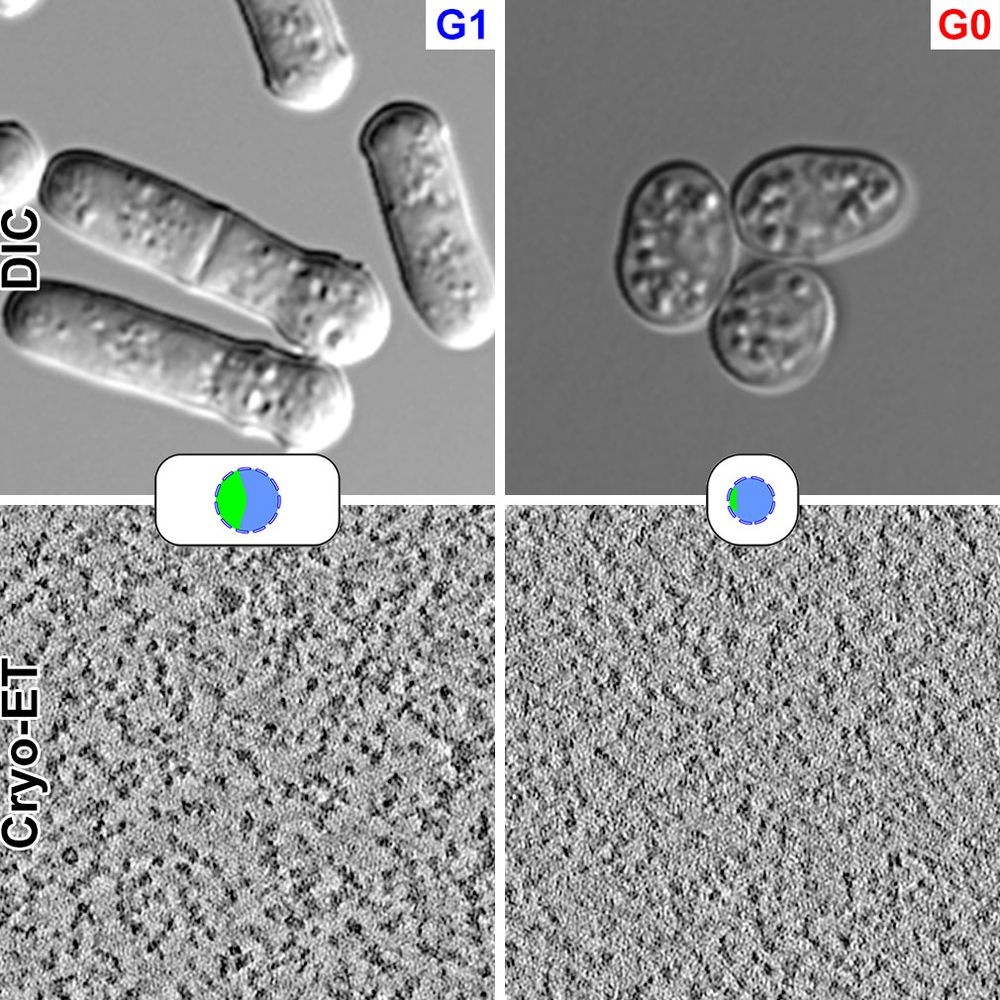
DIC and cryo-ET images of G1 and G0 fission yeast cells.
Zhi Yang Tan & @shujuncai.bsky.social's comparative study of G0 (quiescent) and interphase fission yeast cells is out in @jcellsci.bsky.social doi.org/10.1242/jcs.263654. This study combines multiple cell-biological analyses with in situ cryo-ET. Raw data: www.ebi.ac.uk/empiar/EMPIAR-10339
04.03.2025 17:30 — 👍 21 🔁 12 💬 1 📌 0
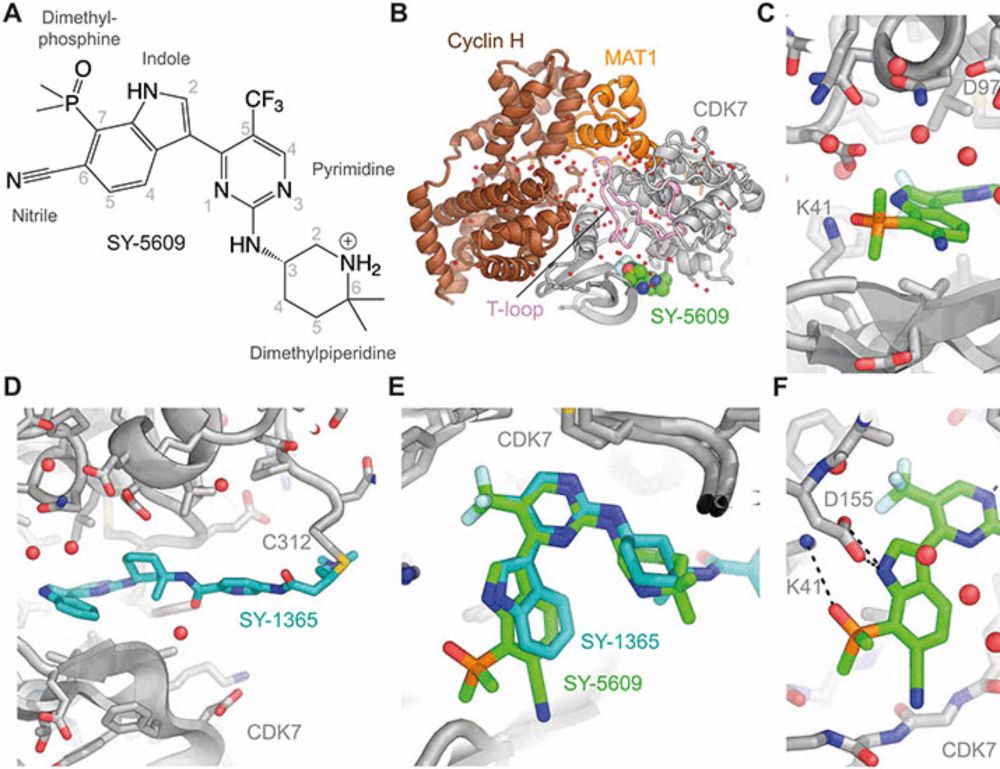
TFIIH kinase CDK7 drives cell proliferation through a common core transcription factor network
CDK7 kinase activity is found to control RNAPII transcription through a cohort of TFs that drive cell cycle and proliferation.
For our latest paper, we worked with SYROS Pharmaceuticals, the groups of Dylan Taatjes and Robin Dowell at CU Boulder, and @abhaykot.bsky.social. Junjie Feng in my lab determined the #cryoEM structure of the CDK7 inhibitor SY5609 bound to its target...
www.science.org/doi/10.1126/...
28.02.2025 21:40 — 👍 58 🔁 19 💬 5 📌 0
Very excited to bring GEARs back with @jonmarkert.bsky.social and @ara-latifkar.bsky.social. We look forward to seeing you in Goldenson 122, Harvard Medical School for our first seminar on 02/27, 6PM.
14.02.2025 03:31 — 👍 5 🔁 3 💬 0 📌 0

Farnung Lab
Excited about mechanistic biology and processes in the cell's nucleus? Join our lab as a post-doctoral researcher! We have a state-of-the-art cryo-EM facility (new microscopes coming!), a brand-new lab space, and a vibrant community at HMS!
DM/email me or check farnunglab.com for more info.
31.01.2025 16:16 — 👍 4 🔁 4 💬 0 📌 1
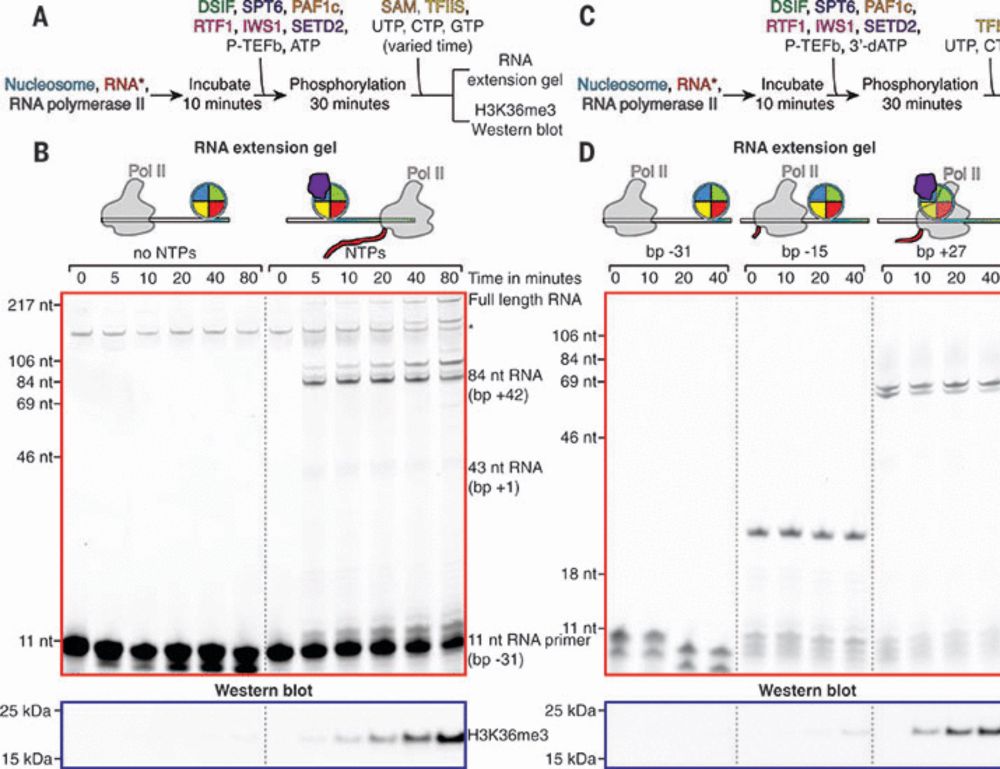
Another highlight from our story: @jonmarkert.bsky.social uncovered how SPT6 binds the exposed H2A–H2B dimer during nucleosomal traversal by Pol II. AlphaFold helped tremendously in validating this interaction. tinyurl.com/setd2 Also check out our H2A–H2B screen: www.biorxiv.org/content/10.1...
18.12.2024 14:08 — 👍 10 🔁 2 💬 0 📌 0
Pol2 stimulates
SETD2 methylation
Of histone H3
#ChromatinHaiku #H3K36
www.science.org/doi/10.1126/...
13.12.2024 15:39 — 👍 20 🔁 4 💬 0 📌 0
Pam taught me so much (how to make nucleosomes, how to use AKTAs…), I’m so incredibly grateful to have worked with her! The entire chromatin field is impacted by her, a very bittersweet day for the Luger lab! I wish her the best!!
13.12.2024 11:43 — 👍 3 🔁 0 💬 0 📌 0
Online today @science.org Structural basis of H3K36 trimethylation by SETD2 during chromatin transcription
by @jonmarkert.bsky.social @lucas.farnunglab.com et al. at @harvardcellbio.bsky.social #cryoEM
#chromatin #transcription #cryoEM #Pol_II🧪🧬❄️🔬
www.science.org/doi/10.1126/...
13.12.2024 02:23 — 👍 31 🔁 4 💬 0 📌 1
Thanks Emma! Wishing you the best in your work at Scripps!
13.12.2024 11:29 — 👍 1 🔁 0 💬 0 📌 0
Thank you CJ! Hope all is well in Madison!
13.12.2024 00:20 — 👍 1 🔁 0 💬 1 📌 0
Assistant Professor at the University of Hong Kong
Dr. Brian Strahl is a chromatin researcher at UNC Chapel Hill studying how epigenetics contributes to human biology and disease. https://strahl-lab.org
PhD student in Narlikar/Ramani labs at UCSF ✨
Group leader @mpi-mg, Berlin
Chromatin tracing, epigenetics, development. Equity and advocacy in academia.
Assistant Prof at Utrecht University, studying the process of transcription elongation vs early termination. (she/her)
Incoming Assistant Professor of MCDB at the University of Michigan. Former JCCF and Leading Edge Postdoc Fellow in the Aaron Whiteley lab at CU Boulder. Predatory bacteria and phage enthusiast obsessed with host-pathogen interactions. She/her.
Sir Henry Wellcome Fellow | Oxford with Ivan Ahel & Harvard with Steve Gygi
Molecular mechanisms of cysteine modifications | GlcNAc and ADP-ribose
Previously: The Francis Crick Institute/Imperial College London (postdoc), University of Dundee (PhD)
Assistant Professor of Biochemistry, Weill Cornell Medicine. www.longlab.org. Studying epigenetics, nucleic acids, stem cells and heart. Cech lab alumnus
Interested in science, RNA, splicing, and single molecule biophysics.
Molecular biologist. Group leader at MRC LMB Cambridge UK. Fellow at Clare Hall college. #CryoEM #RNAbiology #DNArepair
Views/opinions are my own.
https://www2.mrc-lmb.cam.ac.uk/group-leaders/n-to-s/lori-passmore/
Structural biologist studying cilia and ciliopathies.
Alushin lab at Rockefeller University. Interested in cryo-EM, the cytoskeleton, and mechanosensation at the molecular level.
In our lab, hardcore biochemistry meets physics. And coffee. We are investigating DNA replication through single molecule loupe @ Technical University of Munich and MPI of Biochemistry
Mitosis and chromosome biology researcher @rockefelleruniv.bsky.social, New York, NY. Former backstroker. Choir part: bass. Brined turkey & りゅうひ巻き係
funabikilab.com
www.rockefeller.edu/our-scientists/heads-of-laboratories/1169-hironori-funabiki/
PhD candidate in the Farnung lab at Harvard Medical School
Scientist in North Carolina studying chromatin remodelers and transcription factors. @fnucleosome.bsky.social coorganizer
ORCID: 0000-0002-6256-144X
Views and posts are all my own
Bloomberg Distinguished Professor of Structural Biophysics and Chromatin Biology at Johns Hopkins University, #Transcription #DNArepair #Chromatin #cryoEM