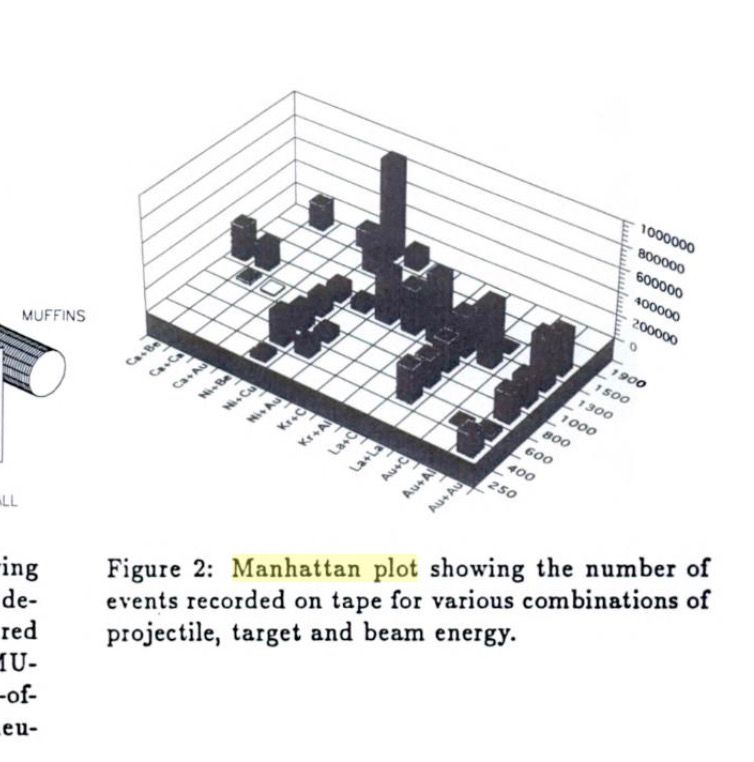
Thanks! This is amazing! So the term 'Manhattan Plot' is not originally a GWAS term at all.
Screenshotting the image here from a 1994 book on nuclear physics, for others who may be interested:
@nikhilmilind.dev.bsky.social
PhD Candidate in the Pritchard Lab at Stanford University. Interested in statistical and population genetics. https://nikhilmilind.dev/

Thanks! This is amazing! So the term 'Manhattan Plot' is not originally a GWAS term at all.
Screenshotting the image here from a 1994 book on nuclear physics, for others who may be interested:
The 2026 Probabilistic Modeling in Genomics (ProbGen) meeting will be held at UC Berkeley, March 25-28, 2026. We have an amazing list of keynote speakers and session chairs:
probgen2026.github.io
Please help spread the news.
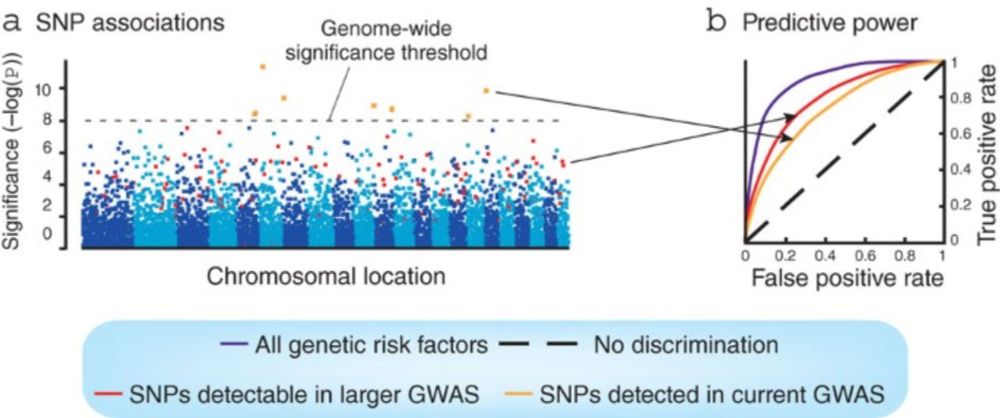
Does anyone know who first used the term "Manhattan Plot"?
Wikipedia links to a 2010 paper by Greg Gibson. I haven't yet found an earlier use, but Greg's phrasing suggests that he did not invent the term.
doi.org/10.1038/ng07...

Can anyone point me to full resolution versions of these classic images from NHGRI that show the growth of GWAS from ~2006--2015? I'm looking for the timelapse versions.
(I know many of you are waiting for the GWAS chapters of my free online textbook on human genetics--they are coming, but slowly!)
I'm thrilled that my lab at NYU is now supported by an NIH MIRA grant! I'm looking to hire 1-2 senior lab members (outstanding postdoc candidates or experienced staff scientists) with expertise in computational or statistical methods in human genetics or genomics. Please share!
25.07.2025 20:29 — 👍 102 🔁 48 💬 4 📌 1My talk from this year's probgen is finally out.
Where do linear mixed models and random effects come from?
They emerge from *mutations* on Ancestral Recombination Graphs.
New paper by Luke O'Connor
@lukeoconnor.bsky.social and Guy Sella @gs2747.bsky.social
Staff scientist position (computational):
I am looking for a computational scientist to join my genomics lab at Stanford. They should have an outstanding skillset in ML/statistical methods for genomic applications, postdoc experience and a strong publication record.
#sciencejobs
Have you worked with human genetics data? Consider taking this short survey to help us better understand how folks are using population descriptors! 🧬💻🧪
26.06.2025 16:15 — 👍 2 🔁 3 💬 0 📌 0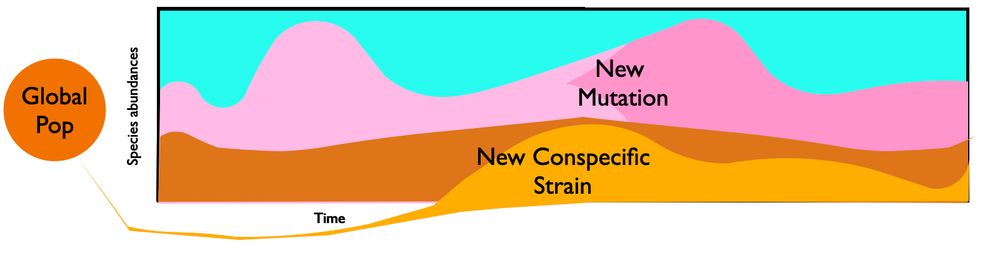
Mueller diagram of ecological and evolutionary dynamics in a gut microbiome. Species fluctuate in abundance and accumulate new mutations. New conspecific strains also colonize from the global population.
Super excited for #Evol2025! I will be sharing some cool work on selection in the gut microbial communities at 9:30 am on Monday in the Evolutionary Ecology I session (Parthenon 2)!! Looking forward to chatting with folks as well!
20.06.2025 16:35 — 👍 22 🔁 6 💬 1 📌 1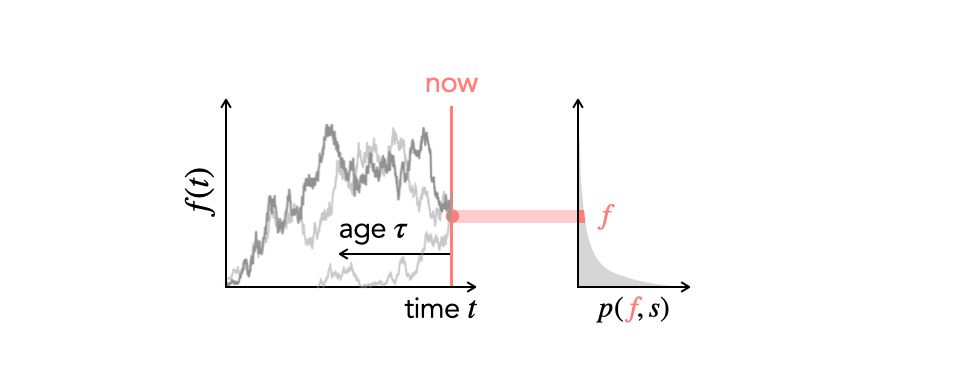
A schematic illustrating the allele frequency trajectories that contribute to a slice of the site frequency spectrum.
Looking forward to #evolution2025! I will be talking about how time-varying demography and selection shape the site frequency spectrum — Saturday at 4:15 pm, Population Genetics Theory IV. Come say hi if you are around!
20.06.2025 16:17 — 👍 20 🔁 4 💬 1 📌 1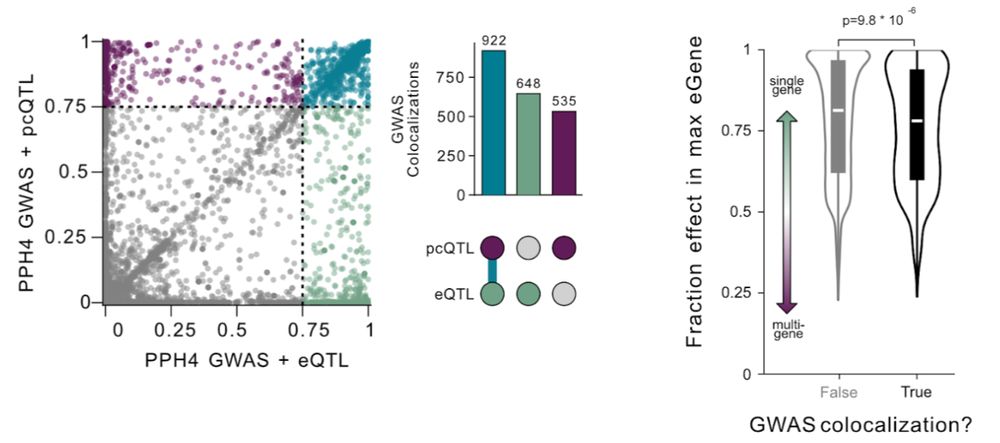
🧵4/5 But are new pcQTLs relevant? We examined associations with GWAS for complex traits and diseases and found over 500 new colocalizations – a 34% increase over single-gene-based analyses! In fact, variants with distributed effects are more likely to be GWAS hits than those affecting just one gene
08.06.2025 17:38 — 👍 5 🔁 2 💬 1 📌 0
Standard methods are equivalent to a flashlight, looking at each gene independently. We combine signals from multiple genes, turning a floodlight onto the genome.
Excited to share my first PhD paper in the @sbmontgom.bsky.social lab with @tamigj.bsky.social (www.biorxiv.org/content/10.1...)! Standard QTL methods treat each gene independently. But what if a single variant regulates multiple nearby genes at once - what we call “allelic proxitropy”? 🧵 ⬇️
08.06.2025 17:38 — 👍 91 🔁 33 💬 6 📌 4Congratulations Jeff! It's been really wonderful to have you in the lab! I cannot wait to see all the fantastic work that will come out of your future lab.
...and trainees, Jeff is an amazing scientist and mentor! If you ever have a chance to work with him you should seize it with both hands!
I'm excited to announce that I'll be starting a lab at UCSF in the @ihgatucsf.bsky.social and @ucsf-epibiostat.bsky.social in July.
We'll work at the intersection of statistical genetics, population genetics, and machine learning.
I have an opportunity to hire a staff scientist for my lab. Looking for someone with outstanding skillset in ML/statistics, genomics applications; interest in mentoring, strong publication record, PD experience required.
Email CV to me+cc my assistant (see 'contact' on my website). Ad to follow.
In any case, it’s useful to plug mechanistic models of how we think an experiment is perturbing the cell into the inputs of deep learning models. This lets us see if our deep learning model has learned a casual relationship that’s consistent with our understanding of the experimental perturbation.
30.05.2025 02:45 — 👍 2 🔁 1 💬 1 📌 0How well can deep learning models predict the effect of modifying chromatin on gene expression???
Our work -- led by Sanjit Batra and Alan Cabrera when they were in @yun-s-song.bsky.social ’s and Isaac Hilton’s labs -- tries to answer this.
🧵🧬🧪
elifesciences.org/reviewed-pre...
Now published in PNAS: www.pnas.org/doi/10.1073/...
23.05.2025 18:39 — 👍 99 🔁 40 💬 1 📌 0Elegant new theory on polygenic adaptation by Will Milligan, Laura Heyward, and Guy Sella.
17.05.2025 23:55 — 👍 56 🔁 26 💬 0 📌 0Hi trainees! I'm at Biology of Genomes and would be thrilled to chat about your work, any questions about academia/science, and Andor. Please don't hesitate to reach out here, by email, or by shouting at me!
#bog25 #bog2025
I hope to see you at ProbGen 2026 on March 25th-28th! I expect a wonderful meeting as always!
07.05.2025 14:57 — 👍 20 🔁 10 💬 0 📌 0I am not sure how to do Bluesky yet, but I want to tell the world about our neighborhood NMF method. First of many collaborations with the Pelka Lab!!
29.04.2025 23:35 — 👍 22 🔁 7 💬 0 📌 0Amidst all the terrible and terrifying news, so lovely to hear of
@jkpritch.bsky.social's election to the National Academy of Sciences. Congratulations!

Congratulations to @jkpritch.bsky.social, very well deserved!
www.nasonline.org/news/2025-na...
Congratulations to the new NAS member @jkpritch.bsky.social. Was very very happy to hear the news! Exceptionally well deserved!
29.04.2025 23:02 — 👍 40 🔁 3 💬 2 📌 0
As a rare bit of good news, a huge congratulations to @jkpritch.bsky.social who was just elected to the National Academy of Sciences.
Here's the full list.

A dragon emerges from an egg, within a nest among others waiting to hatch atop a majestic castle. The dragon is embedded with a DragonRNA, a DNA strand covalently extended with an RNA tail. Variants of precursor DNAs are backlit by a glowing polymerase within each egg,
28.04.2025 21:18 — 👍 2 🔁 2 💬 1 📌 0
What is epistasis?
New preprint Blue-torial!
(Work by the wonderful postdocs Maryn Carlson and Bryan Andrews)
www.biorxiv.org/content/10.1...
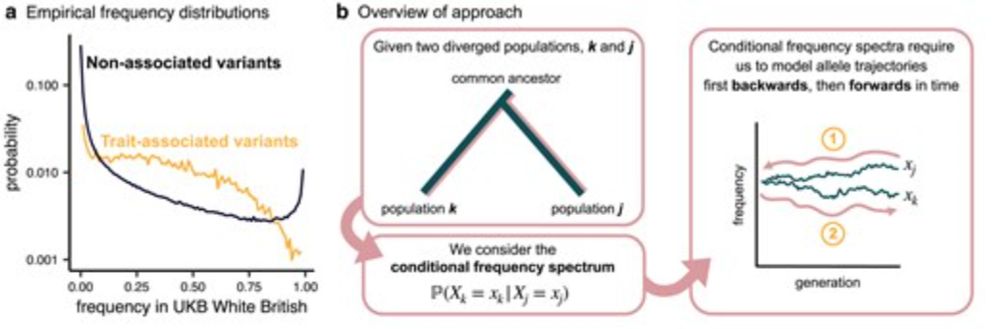
a quick note on my paper with @jeffspence.bsky.social and @jkpritch.bsky.social on conditional frequency spectra, now out in a @genetics-gsa.bsky.social special issue: doi.org/10.1093/genetics/iyae210
18.04.2025 19:49 — 👍 61 🔁 25 💬 3 📌 1