As a big fan and sometimes baker of science-themed cakes, this is amazing Rosie! 😍 Perfect way to honour an exciting Nobel Prize!
09.10.2025 22:23 — 👍 4 🔁 0 💬 1 📌 0

Seminar time⏲️ We welcome Professor Dimitrios Fotiadis, IBMM, Medical Faculty, University of Bern: “Structural and mechanistic insights into transmembrane glucose transport and light-driven proton pumping” 🥳More info: ccemmp.org/events/ccemm...
11.08.2025 03:13 — 👍 1 🔁 1 💬 0 📌 0

Join our Cryo-ET workshop and learn sample preparation, grid screening and data collection.
📍Ian Holmes Imaging Centre
🗓️9th-10th September 2025
🔗Register now: rduevents.unimelb.edu.au/event/2271-c...
#CryoET #ElectronMicroscopy #StructuralBiology #CCeMMP
11.07.2025 04:54 — 👍 9 🔁 4 💬 0 📌 0

An image of a large diamond crystal structure model with a large sign on the top of it

A group of people sat around the giant diamond structure

detail of someone putting together the giant diamond structure
Many in the #Crystallography community will remember this (and may have helped put part of it together). Our record breaking diamond structure built at the 2023 IUCr congress in Melbourne
[Photo credit Jun Aishima]
02.06.2025 04:17 — 👍 9 🔁 6 💬 1 📌 1
Congratulations Javaid!
06.06.2025 09:10 — 👍 0 🔁 0 💬 1 📌 0
Cryo-EM was conducted at Ian Holmes Imaging Centre at, with data collected by Hamish Brown, with contributions from
@bio21director.bsky.social , @bigfrenchman.bsky.social
, Michelle Christie, Riya Joesph, Craig Morton and Rod Tweten and team. #cryoEM
30.03.2025 10:53 — 👍 1 🔁 0 💬 0 📌 0
This work was part of my PhD at UniMelb and was my first foray into cryo-EM! Admittedly, learning the various steps of cryo-EM on proteoliposome samples felt like jumping in the deep end so I am very happy and proud for all to see how it worked out.
30.03.2025 10:50 — 👍 1 🔁 0 💬 1 📌 0

These structures reveal how multiple regulatory features, such as proteolytic activation, ensure pore formation. Furthermore, we find features that are not common to the related CDC family of pore-forming toxins. Instead, CDCLs seem show features that straddle the CDC, MACPF and gasdermin families.
30.03.2025 10:50 — 👍 0 🔁 0 💬 1 📌 0

This includes the crystal structure of the proteolytically-activated form and cryo-EM structures of the inserted pore and prepore-like complex, solved on the surface of liposomes.
30.03.2025 10:50 — 👍 2 🔁 0 💬 1 📌 0
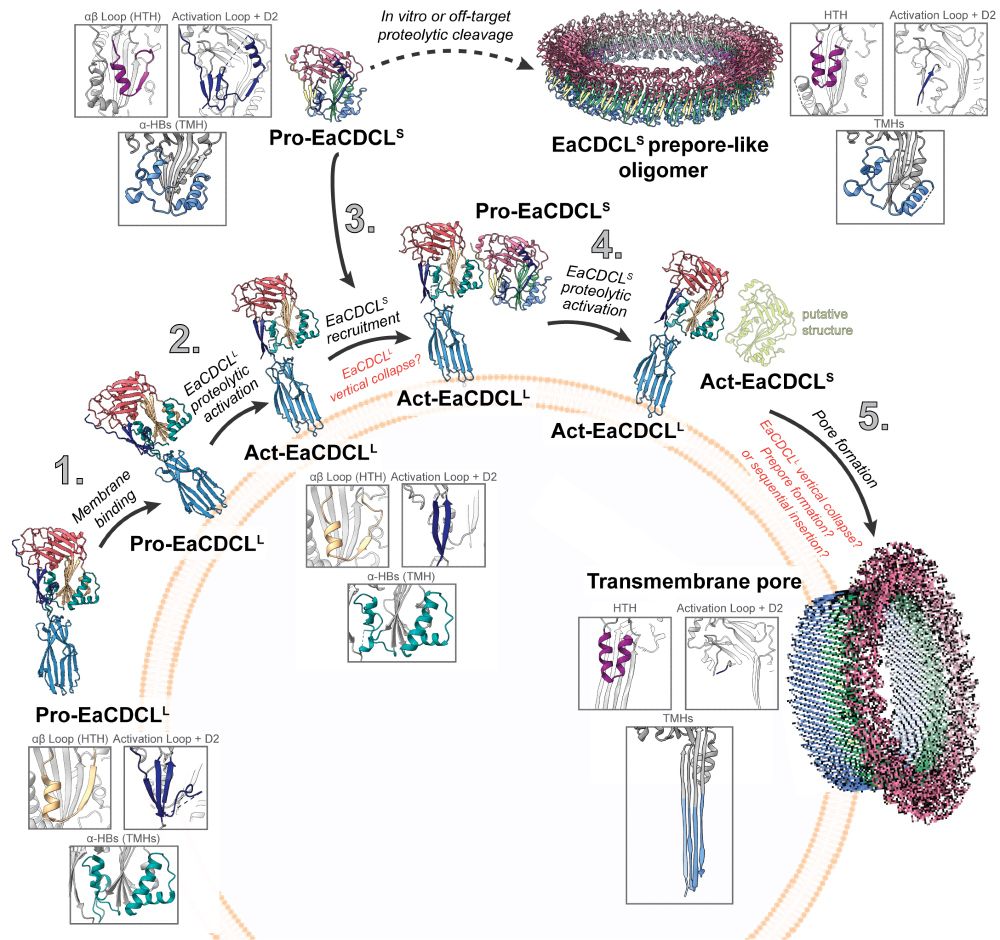
Excited to share that our work on a family of pore-forming proteins is now live in Science Advances! We show structural snapshots across the entire pore-forming pathway for a cholesterol-dependent cytolysin-like (CDCL) bicomponent system.
30.03.2025 10:50 — 👍 32 🔁 9 💬 3 📌 0
Our paper on the structure and evolution of baculovirus is out! Excited to share work I did at Monash in the Coulibaly lab, with big shout outs to co-authors Josh and Jungmin, collaborators Mart and team and everyone else who contributed. Definitely one of the coolest structures I'll ever work on 🤩
24.12.2024 01:16 — 👍 23 🔁 4 💬 0 📌 0
Postdoc in the Grinter Lab at Bio21, UoM
Antibiotic mechanisms| Cryo-EM | Bacterial pathogens | Membrane proteins
Professor @ University of Sydney; interested in membrane transport proteins, biophysics, pharmacology and diversity and inclusion in STEMM. Views expressed here are my own. She/her.
PhD student in structural biology with @greening.bsky.social and @knottrna.bsky.social at Monash Uni. (he/him)
Interested in hydrogenases, evolution, protein design.
💻 https://www.jameslingford.com/
Virologist @UCPH_Research | Assistant Professor @Prentoe_Lab | Interested in viral escape and envelope proteins structures 🦠
Bioinformatics, protein modeling, cryoEM, drug screening, function prediction. Daisuke Kihara, professor of Biol/CS, Purdue U. https://kiharalab.org/ YouTube: http://alturl.com/gxvah
Prof @sydneyuni.bsky.social Engineer and scientist building bridges between research & industry at USYD & @ansto.bsky.social, Australia.
She/Her. Mum & wife. Leader & Mentor. Loving ⛷🏄♀️ & life in general! Comments are my own
Structural virology / Structure-based vaccine design
mclellanlab.org
Professor of Molecular Biosciences
The University of Texas at Austin
Opinions are my own and not the views of my employer
*Unattended account*. Instantly reposts posts from @arc-tracker.bsky.social's bot. That is, this account should only post announcements of ARC grants & recommendations to the Minister. See pinned thread (esp. 2nd post) for how to get device notifications.
MX Beamline Scientist at Australian Synchrotron. Crystals, chemistry and cooking.
She/her
We're a public-spirited institution making distinctive contributions to society in research, learning, teaching and engagement.
CRICOS code: 00116K
CryoEM of bacterial filaments and eukaryotic cytoskeleton. Postdoc within the Ghosal Lab at University of Melbourne, Australia. Formally at Birkbeck, University of Bristol.
Cell Press partners with scientists across all disciplines to publish and share work that will inspire future directions in research. #ScienceThatInspires
Briggs group at MPI Biochemistry
Department of Cell and Virus Structure
Cryo-EM, tomography, CLEM, coated vesicles, enveloped viruses.
Structural Biologist. Cryo-EM. Mitochondria and Neurodegeneration. PhD at the Bio21 institute at the University of Melbourne.
🔬 We research and develop treatments for cancer, diseases of development and ageing, immune disorders and infections.
https://www.wehi.edu.au
Doctoral candidate at MDC Berlin focusing on Structural Biology | Exploring membrane protein complexes via in-situ Cryo-ET ❄️
Framework materials, total scattering & Fourier transform enthusiast │ Curator of patterns and alternative crystallographic teaching at behance.net/specialdefects
https://orcid.org/0000-0002-6817-0810
A group of pattern scientists (crystallographers) sharing posts using the #BraggYourPattern hashtag, see a pattern, take a photo, share and enjoy! https://braggyourpattern.com/
Follow for the crystal structures, stay for the cricket (or vice versa)
Interests brought to you by the letter ‘C’, cryominerals 🥶, woman’s cricket 🏏 and cookbooks 🥘
Lives on Dharug and Gandangarra country (Blue Mountains, Australia)
Ph.D Student @Schuller Lab @Uni_Marburg | Structural Biologist | McGill University Alumnus | #Photosynthesis 🌱 #CryoEM/ET❄🔬 #Biochemistry🧬 🇮🇳->🇨🇦->🇩🇪







