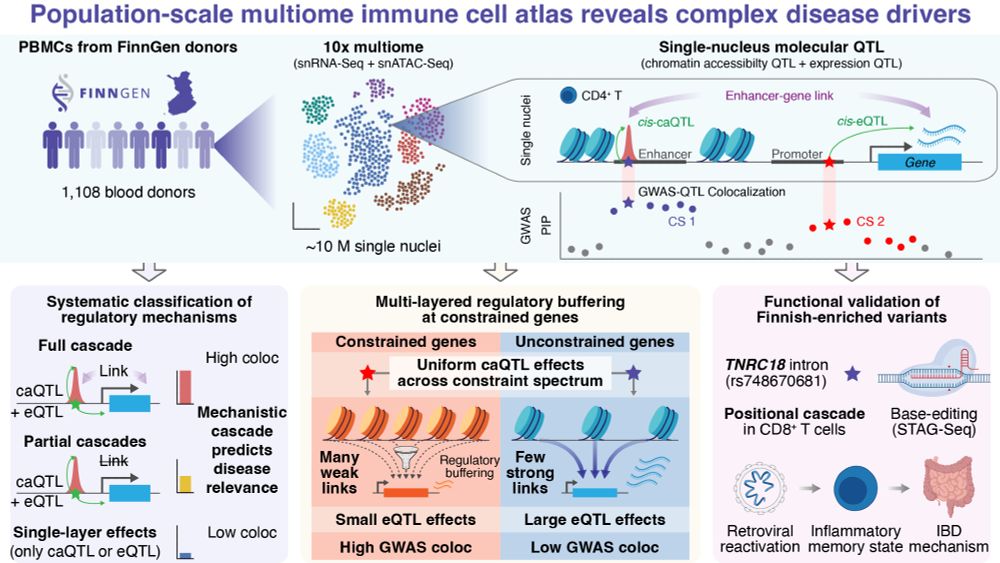
Excited to share our new FinnGen single-nucleus multiome preprint! 🧬
We profiled ~10M PBMCs (snRNA-seq + snATAC-seq) from 1,108 Finnish donors to map how genetic variants drive complex disease through chromatin and gene regulation 🧵👇
🔗 Link: www.medrxiv.org/content/10.1...
01.12.2025 15:36 — 👍 33 🔁 19 💬 1 📌 0
If you’re looking for highly heritable quantitative traits (beyond height!) to test your new methods, these “lab values” could be your next model traits!!
We did so in our pedigree-BLUP work:
we estimated family-based h2 & compared it to SNP h2 to clarify the different concepts of heritability!!
24.10.2025 01:39 — 👍 9 🔁 0 💬 0 📌 0
New from FinnGen!! 🚀🚀🚀
FinnGen just released GWAS summary statistics for 383 lab measurements (OMOPIDs) - each based on ≥1,000 participants!!
All results are open for browsing & download:
👉 labvalues.finngen.fi.
🧬🧬🧬
Have fun!! 🏄🏄♀️🏄♂️
24.10.2025 00:09 — 👍 12 🔁 1 💬 0 📌 1
A traditional "Comment" typically wouldn’t include such a "complicated" figure - we have to follow journal guidelines (=delete it from the paper).
However, we’re free to share the full version with you here!!
For those who think in pictures (myself included), I hope you find it clear & helpful! 🫶
17.10.2025 02:43 — 👍 1 🔁 0 💬 0 📌 0
Excited (😁) to share a commentary on @finngen.bsky.social & cancer research, co-written with Mervi Aavikko & @dalygene.bsky.social!!
(View-only full text -> rdcu.be/eLlJs)
16.10.2025 22:27 — 👍 7 🔁 1 💬 1 📌 0
Join Jesse & me for a 🌟 #ASHG2025 Featured Symposium!!
"Decoding Human Aging: From Single-Cell Resolution to Population-Scale Insights"
⏰ Oct 17, 8:30-10:00 AM
🏡 258ABC, Level 2
With a fantastic ✨ speaker lineup - @albarmeira.bsky.social & Albert (Kejun) & Giulio & @austinargen.bsky.social!
2/n
14.10.2025 15:45 — 👍 6 🔁 0 💬 0 📌 1

Happy (😀) to invite you to our Talk/Featured Symposium/Poster/Booth at #ASHG2025!!
⏰ Oct 15, 1:30 -1:45 PM
🏡 258ABC, Level 2
I'll be giving a talk on sex chromosome trisomies (XXY, XYY, XXX) - and the surprising new biology of chromosomes X and Y!!
Link: meetings.ashg.org/event/ASHG25...
1/n
14.10.2025 15:35 — 👍 6 🔁 0 💬 0 📌 1

Meet FinnGen at the ASHG 2025! | FinnGen
Results based on the FinnGen data are presented in almost 40 talks or posters during the 2025 Annual Meeting of the American Society of Human Genetics (ASHG) in Boston, October 14-18. We also have a b...
We’re getting ready for an exciting week at #ASHG2025 in Boston! Come meet the FinnGen team at booth 147 - we’ll be sharing updates and showcasing new public resources and tools. Plus, don’t miss the various presentations featuring FinnGen results across the program!
www.finngen.fi/en/meet-finn...
10.10.2025 11:59 — 👍 4 🔁 3 💬 0 📌 1

We are excited to share GPN-Star, a cost-effective, biologically grounded genomic language modeling framework that achieves state-of-the-art performance across a wide range of variant effect prediction tasks relevant to human genetics.
www.biorxiv.org/content/10.1...
(1/n)
22.09.2025 05:29 — 👍 175 🔁 90 💬 4 📌 5
Thanks, Cristian!! 😄😄😄
21.08.2025 16:39 — 👍 0 🔁 0 💬 0 📌 0
Our trisomy work is now online! 🚀
Got curious after reading our short PheWAS paper? 👀
Catch me 🌠 & my talk 🎤 this October at #ASHG25 in Boston!
Yes, I am still endlessly curious about sex chromosome aneuploidies - whether it's somatic (X/Y loss) or germline (trisomies)!! 💘
20.08.2025 23:25 — 👍 17 🔁 2 💬 1 📌 0
And Figure 1 in the preview summarizes everything so nicely - way better than we authors could!!
Preview: www.cell.com/cell-genomic...
Original research article: www.cell.com/cell-genomic...
11.06.2025 22:05 — 👍 2 🔁 0 💬 0 📌 0
And, the figure one you generated is so informative!!
11.06.2025 21:46 — 👍 2 🔁 0 💬 1 📌 0
Thanks @struangrant.bsky.social and @agacita.bsky.social for seeing the uniqueness of our work and guiding others to understand it better!! Really appreciate your vision and insight!!
11.06.2025 21:18 — 👍 2 🔁 0 💬 1 📌 0
Wow!! Big thanks for the very lovely preview!! 🫶🫶🫶
TOO MANY analyses -> very DIFFERENT feedback on (several of) our submissions - some thought it was too lengthy, others considered the design creative, rigorous.
Perhaps no need to do standard analyses if the data/question itself is not standard...
11.06.2025 21:14 — 👍 3 🔁 1 💬 0 📌 1
Thanks to the @finngen.bsky.social team for years of effort 🫶 in making it a unique resource for genetic discovery, and congratulations to FinnGen researchers (including us 😉) for utilizing the resources smartly!!
26.05.2025 15:52 — 👍 3 🔁 0 💬 0 📌 0

Hi - Excited to share our two talks at @eshg.bsky.social about @finngen.bsky.social!
Because of ... I cannot join in person, but I will answer your questions online (or write to me)! 🧬
Feiyi & Zhiyu are in Milan now; discuss the BLUP work with them (if you can catch them❤️🔥)!
Enjoy the conference!
24.05.2025 13:31 — 👍 4 🔁 1 💬 0 📌 0
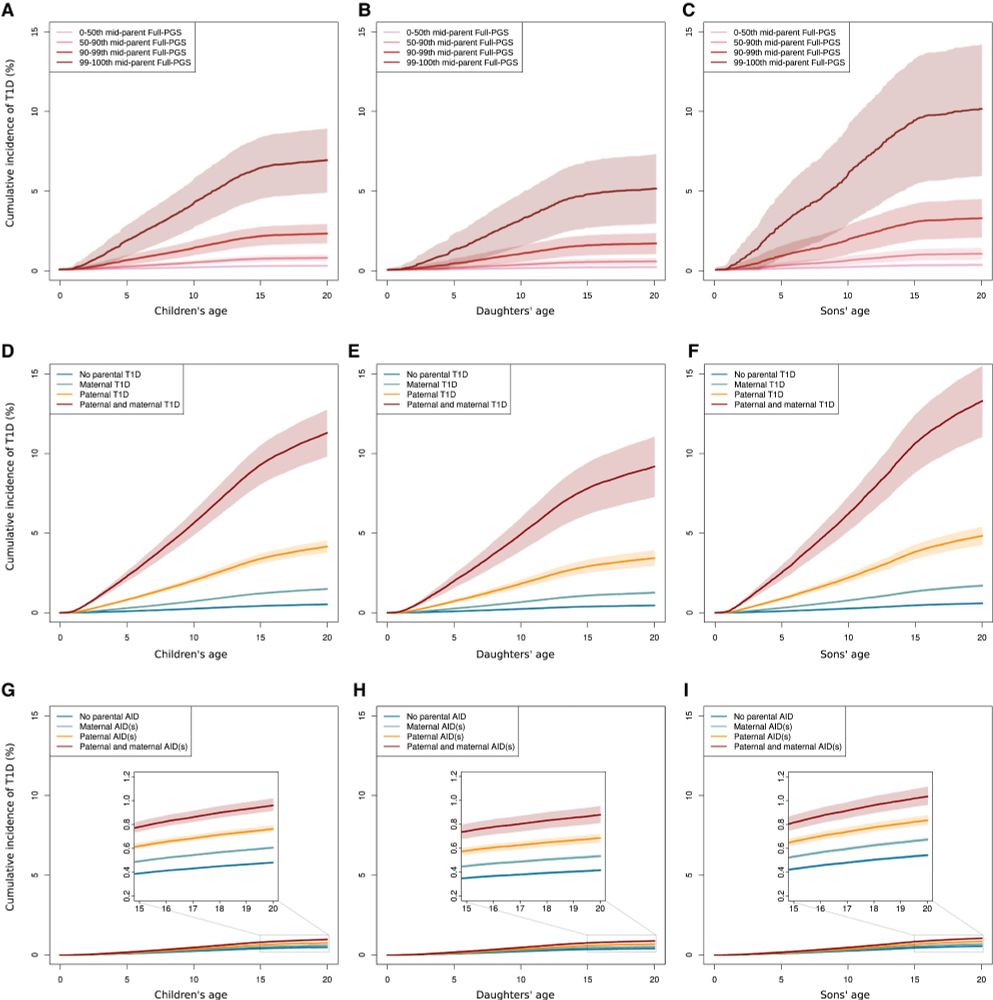
8/n
Parental T1D PGS is a strong predictor of T1D risk in children.
About parental PGSs of other AIDs - not adding too much to the prediction, given the high accuracy of T1D PGS.
PGS vs family history? High parental PGSs are usually more informative than a diagnosis of T1D in the mother or father.
02.05.2025 17:36 — 👍 1 🔁 0 💬 0 📌 0

7/n
People with AIDs are less likely to have children - for many reasons, including fear of transmitting diseases to offspring.
By estimating T1D risk for their children, couples can have informed family planning.
Also, children at high risk can benefit from targeted screening & early prevention.
02.05.2025 17:07 — 👍 1 🔁 0 💬 1 📌 0

6/n
With pTDT, we see 2 main patterns for parent AID -> offspring T1D inheritance:
Pattern 1 – Overtransmission of both HLA & non-HLA PGSs for parental T1D, HYPO, RA, SLE.
Pattern 2 – Transmission occurs only for HLA: overtransmission for parental CD & psoriasis, undertransmission for IBD & MS.
02.05.2025 16:29 — 👍 1 🔁 0 💬 1 📌 0

5/n
But our original curiosity is about parents & children...
We can run polygenic transmission disequilibrium tests (pTDT) with AID HLA & non-HLA PGS in FinnGen trios!
A reminder - pTDT checks if AID risk variants are passed on more or less to children with T1D compared to what would be expected.
02.05.2025 15:55 — 👍 1 🔁 0 💬 1 📌 0
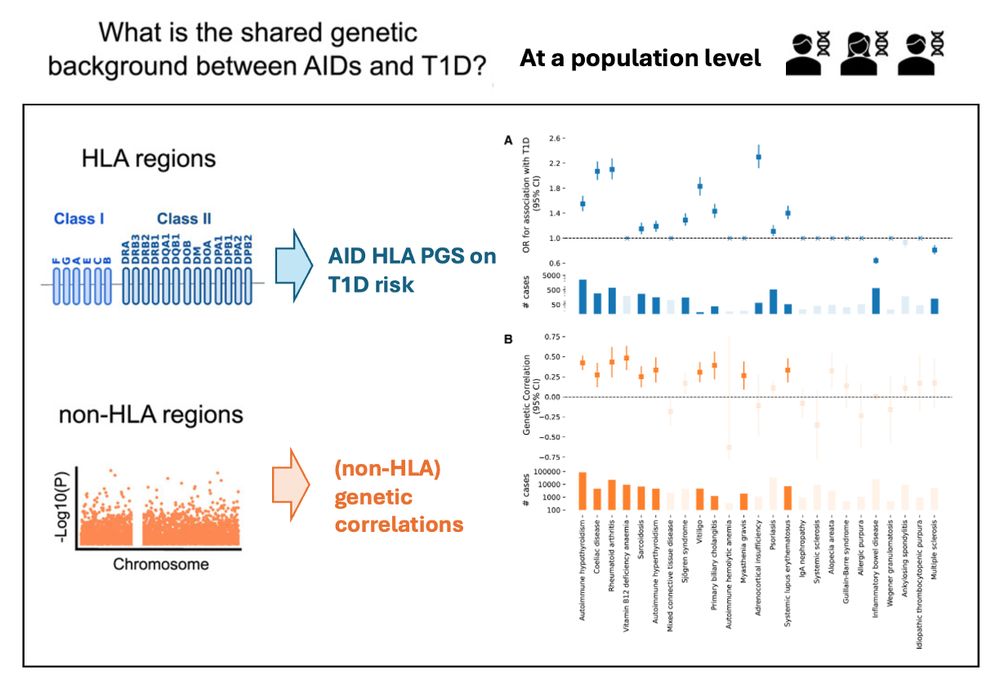
4/n
Now, let's revisit the question of the shared genetic background! 🧬🧬🧬
Overall, genetic analyses are consistent with epidemiological findings. For example, for HYPO, CD, RA, both HLA & non-HLA show positive effects.
However, for MS & IBD, non-HLA has no effect, while HLA has protective effects.
02.05.2025 15:21 — 👍 1 🔁 0 💬 1 📌 0

3/n
Genetic correlation is straightforward but remember HLA - largely impacting T1D & other AIDs - is excluded by LDSC.
If genetic correlation is a genome-wide non-HLA measurement, how can we get HLA back into the game?
We built a within-sample HLA PGS with weighted ridge classifiers in FinnGen.
28.04.2025 03:15 — 👍 6 🔁 1 💬 1 📌 0

2/n
To what extent do the identified epidemiological associations result from genetics?🧬🧬🧬
Let’s download some summary stats and estimate some genetic correlations (quickly).
It looks not too bad - if we put the plot of epidemiological analysis aside for comparisons.
28.04.2025 02:33 — 👍 3 🔁 0 💬 1 📌 0

1/n
Which parental autoimmune diseases (AIDs) are associated with T1D in offspring?
With 14,571 trios having T1D-affected children and 43,713 matched control trios (1:3) in Finnish nationwide registers FinRegistry, we identified 9 AIDs associated with increased risk of T1D in offspring.
28.04.2025 01:22 — 👍 2 🔁 0 💬 1 📌 0
Why @finngen.bsky.social & why T1D?
1⃣ Finland has the world’s highest incidence of T1D.
2⃣ National healthcare data has been collected for decades -> disease status available for 2 generations.
3⃣ FinnGen genotyped 1/10 of the population + family-based legacy cohorts -> 1.2K T1D genotyped trios.
26.04.2025 21:59 — 👍 3 🔁 0 💬 1 📌 0
Join us April 8-9 for @geneticssociety.bsky.social Spring Symposium on Cancer Genetics!! 🧬
I will present our work on #mCAs & #solid_tumors in @finngen.bsky.social.
Register link: learning.ashg.org/products/202...
Excited to connect with you virtually months before the Boston annual meeting!! 🫶
25.03.2025 22:47 — 👍 5 🔁 1 💬 0 📌 0
Computational Biologist in the Narita Lab, CRUK CI and CRA at Homerton College, University of Cambridge. 🩷 🐈🐈⬛🧶🧮💻🧬🧪🧫🔭
Cancer researcher at Hunter College of the City University of New York & Weill Cornell Medicine. Views are my own.
Assistant Professor @YaleMed | tissue biology, computation, systems biology |
www.hattiechunglab.bio
Director of Finance, Membership and Publishing at the British Society for Immunology (BSI), also Treasurer/Trustee of Applied Microbiology International (AMI). Dad who loves science and sports (especially #immunology and #taekwondo) All views are my own.
Executive Publisher at T&F. Publish journals. Walk dogs. ❤️ doughnuts & ice cream. #scicomm #physicalsciences #mathematics #statistics #datascience #history #science #STS
PhD Candidate in the Pritchard Lab at Stanford University. Interested in statistical and population genetics.
https://nikhilmilind.dev/
Genetics of Perinatal and Reproductive outcomes https://mosaic-wlab.github.io/
PhD student of Human Genetics at McGill. Studied genomics at LCG UNAM. Single cell, statistics, cancer.
Professor of Medicine at Vanderbilt University Medical Center and Director of the Vanderbilt Genetics Institute. My research is in computational and statistical human genomics.
JupyterLab is my Winamp visualizer.
Researcher in statistical genetics looking for new professional opportunities
PhD from Oxford
lfe.pt
Genetics and Genomics, Computational Biology
Crossing disciplinary borders of psychology, social sciences, and molecular genetics to answer why people do good and feel well. Life course, intergenerational transmission, gene-environment interactions, siblings. Finnish genetically-informed registers
Professor of Molecular Epidemiology in Cambridge (UK)
Lead Research Bioinformatician at Genomics England.
Incoming DDLS Assistant Professor, Data Science and AI
division, Chalmers University | Formerly at JHU-LangmeadLab, UNIL-DessimozLab, WUR-deRidderGroup | Pangenomics for studying genome variation and evolution | Hiring PhDs/Postdocs CGRLab.github.io
Postdoctoral Research Fellow, DBMI, HMS, Neurology, BWH
Sudarsky Scholar'22
MIT EECS SM'14, PhD'19
Fulbright S&T Fellow'12
Ludwig Center Fellow'15
She/Her/Hers
Research Assistant Professor within the Division of Bioinnovation and Genome Sciences at the Translational Genomics Research Institute, part of City of Hope. heinin.github.io
Postdoc at Siepel Lab, CSHL
Statistical genetics, phylogenetics, and optimization














