Thanks Leo! It was lovely to meet you again!
02.07.2025 17:04 — 👍 2 🔁 0 💬 0 📌 0Hyejung Won
@hyejungwon.bsky.social
Associate Professor at UNC Chapel Hill Neurogenetics wonlab.org
@hyejungwon.bsky.social
Associate Professor at UNC Chapel Hill Neurogenetics wonlab.org
Thanks Leo! It was lovely to meet you again!
02.07.2025 17:04 — 👍 2 🔁 0 💬 0 📌 0Thanks so much Keri for inviting me ❤️ It was a great pleasure to visit Lieber and meet so many amazing scientists!
02.07.2025 17:03 — 👍 1 🔁 0 💬 0 📌 0
Phenomenal talk from @hyejungwon.bsky.social who is visiting us @lieberinstitute.bsky.social for the day. Groundbreaking work w/ important implications and roadmap for using rapidly emerging information on psychiatric genetics to prioritize variants and understand their impact on 🧠 function
24.06.2025 19:23 — 👍 11 🔁 3 💬 2 📌 0Our "Atlas of Variant Effects 2030 Roadmap" is live: zenodo.org/records/1542...
1/n
How does APOE change your risk of developing Alzheimer's disease? At least partially by changing the function of immune cells in your brain 🧠🧬
New paper out in @natcomms.nature.com today: www.nature.com/articles/s41...
@ukdri.ac.uk @kingsioppn.bsky.social @imperialbrains.bsky.social
Huge congrats Debby! This was a lovely story when you presented at GRC ❤️
14.05.2025 21:30 — 👍 4 🔁 0 💬 1 📌 0Huge congrats Alexi and Sarah 🎉
01.05.2025 13:16 — 👍 2 🔁 0 💬 1 📌 0Silencing of transgenes during iPSC differentiation is a frequent problem. 🧬🫢
Check out the piggyBac system below, which has resisted silencing in many protocols across many labs. 💪
Excited that our lab could contribute to this team effort led by @tuenaka.bsky.social & Marius Wernig!
Watch the video📽️attached to see what each of these awesome #FemsinSTEM are up to!
🧵3/3
Marija, I am so much happy for you! Huge congrats! 😍
06.03.2025 22:15 — 👍 1 🔁 0 💬 1 📌 0
Hello, it is because borderline disorder GWAS is not well powered: i.e. it does not have any hits (dots) above the red line in figure 2: www.nature.com/articles/tp2.... This means that there are not robust and reliable genomic regions that we can chase after for functional analysis.
28.02.2025 21:49 — 👍 1 🔁 0 💬 0 📌 0This is soooo cool Pedro! Huge congrats!
27.02.2025 15:49 — 👍 0 🔁 0 💬 1 📌 0
Very good new preprint by @nanamatoba.bsky.social @hyejungwon.bsky.social @steinlab.bsky.social, using a Massively Parallel Reporter Assay to gain insight into regulatory elements associated with inter-individual differences in cortical structure (Wnt response sensitivity plays a prominent role)🧪🧠🧬
12.02.2025 09:44 — 👍 6 🔁 2 💬 0 📌 015/ Last but not least, I'd like to thank fabulous first authors, Sool Lee, Jessica McAfee, and Jiseok Lee; great collaborators, Pat Sullivan, Adriana Beltran, @markgerstein.bsky.social, Alan Boyle; and a fantastic editor, Scott Behie!
28.01.2025 14:45 — 👍 2 🔁 0 💬 0 📌 0Thanks so much!
28.01.2025 14:34 — 👍 1 🔁 0 💬 1 📌 0Yay thanks! You deserve a credit here 😆
28.01.2025 14:34 — 👍 1 🔁 0 💬 0 📌 014/ There’s much more in the full manuscript, including findings on Alu elements with gene regulatory activity and CRISPR validation. Check it out here for full access: authors.elsevier.com/a/1kU9ZL7PXq...
28.01.2025 14:30 — 👍 1 🔁 0 💬 1 📌 013/ Together, our findings suggest pleiotropy arises from the functional properties of target genes and proteins, including prolonged gene activity during neuronal differentiation and higher protein connectivity in PPI networks.
28.01.2025 14:30 — 👍 1 🔁 0 💬 1 📌 012/ PPI networks were a recurring theme: mDis3-bound transcription factors, mDis3 proteins, and even downstream effectors all showed higher connectivity in PPI networks. Connectivity in these networks seems critical to how pleiotropic variants exert their effects.
28.01.2025 14:30 — 👍 0 🔁 0 💬 1 📌 0
11/ We also investigated protein-protein interaction (PPI) networks. mDis3 proteins were significantly more interconnected than mDis1 proteins. This implies pleiotropic proteins may amplify their effects through network connectivity, rippling through molecular pathways.
28.01.2025 14:30 — 👍 0 🔁 0 💬 1 📌 010/ This suggests that mDis3 variants target genes with broader or longer activity across excitatory neuronal differentiation, potentially driving multiple downstream effects. These temporal differences may explain part of the functional diversity seen in pleiotropy.
28.01.2025 14:30 — 👍 1 🔁 0 💬 1 📌 09/ We next examined the functional properties of variants and their target genes. Both mDis1 and mDis3 variants were enriched in the excitatory neuronal lineage. In this lineage, mDis1 genes were often expressed in a single cell type, while mDis3 genes showed prolonged, multi-cell-type expression.
28.01.2025 14:29 — 👍 0 🔁 0 💬 1 📌 0
8/ Next, we wondered if mDis3 variants have more complex regulatory connectivity (e.g., linking to multiple genes). Again, no significant differences were found between mDis3 and mDis1 loci in the number of genes connected to emVars.
28.01.2025 14:29 — 👍 0 🔁 0 💬 1 📌 07/ First, we asked if mDis3 loci simply have more emVars than mDis1 loci. The number of emVars didn’t differ between mDis3 and mDis1 loci. So, the extent of pleiotropy wasn’t explained by the number of variants with regulatory activity.
28.01.2025 14:28 — 👍 0 🔁 0 💬 1 📌 06/ Why exclude 2-disorder variants? Many were linked to bipolar and schizophrenia, which have high genetic correlation (>0.7). This made it hard to determine if they were truly pleiotropic or just driven by shared biology between these two highly related disorders.
27.01.2025 19:03 — 👍 1 🔁 0 💬 1 📌 0
5/ MPRA narrowed ~17k variants to ~680 with validated regulatory activity, which we call expression-modulating variants (emVars). We categorized emVars into: (1) mDis3: linked to 3+ disorders. (2) mDis1: linked to 1 disorder. Variants linked to 2 disorders were excluded.
27.01.2025 19:03 — 👍 2 🔁 0 💬 1 📌 0
4/ One challenge: ~17k variants in cross-disorder loci showed nominal associations (p < 1e-5), making it hard to pinpoint key variants. To address this, we used MPRA (massively parallel reporter assay) to test the regulatory activity of thousands of variants simultaneously.
27.01.2025 19:03 — 👍 1 🔁 0 💬 1 📌 0
3/ In 2019, the PGC cross-disorder group conducted a meta-GWAS of 8 psychiatric disorders, identifying 136 “cross-disorder loci.” Among these, 109 loci were linked to 2+ disorders (pleiotropic loci). But how do pleiotropic loci differ from disorder-specific loci?
27.01.2025 19:02 — 👍 1 🔁 0 💬 1 📌 0
2/ Pleiotropy occurs when genetic variants affect multiple traits. It's common in psychiatry, where many disorders show significant genetic correlations.
27.01.2025 19:02 — 👍 1 🔁 0 💬 1 📌 0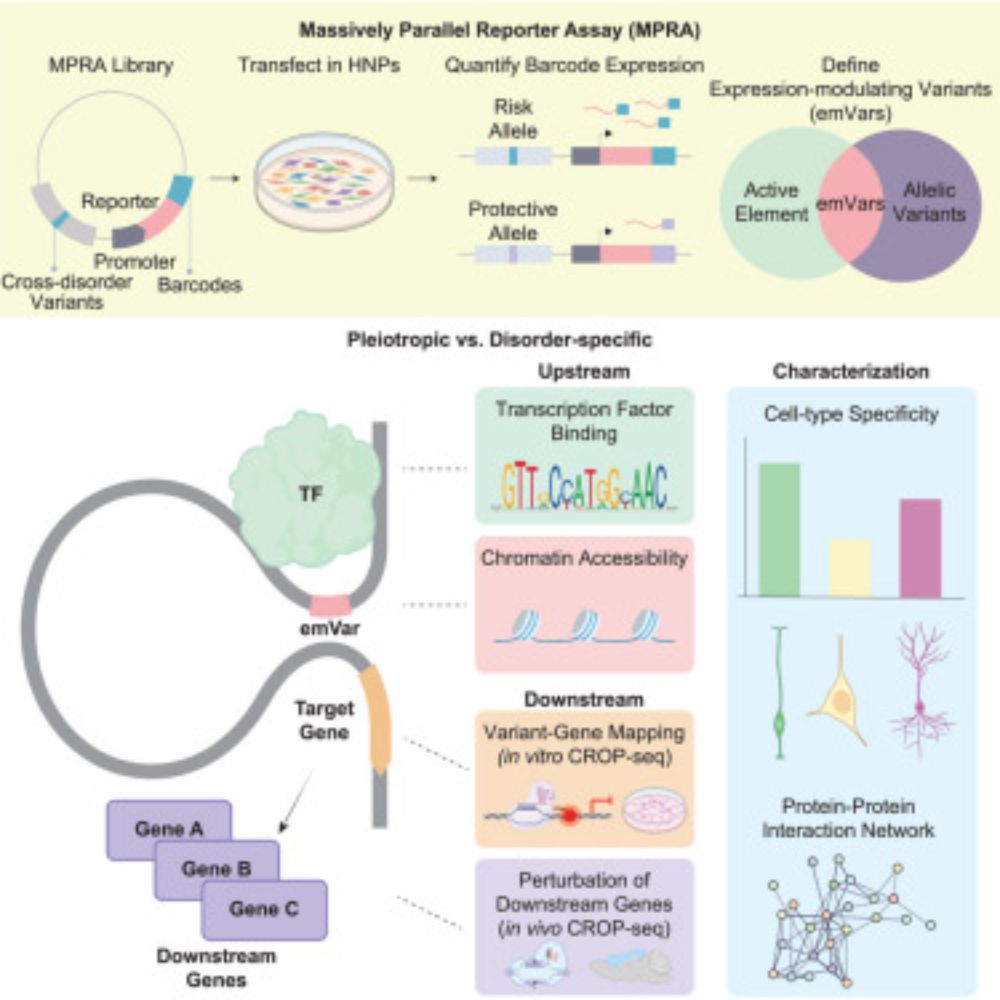
1/ I am beyond thrilled to share our new paper in @CellCellPress! We explored the regulatory principles underlying pleiotropy in psychiatric disorders.
www.cell.com/cell/abstrac...