🙌 Huge thanks to the gnomAD team & collaborators — this is a step toward more equitable & clinically useful population frequency resources.
06.10.2025 18:45 — 👍 0 🔁 0 💬 0 📌 0🙌 Huge thanks to the gnomAD team & collaborators — this is a step toward more equitable & clinically useful population frequency resources.
06.10.2025 18:45 — 👍 0 🔁 0 💬 0 📌 0👆The gnomAD browser is now up and running with LAI data live for both genetic ancestry groups!
06.10.2025 18:45 — 👍 0 🔁 0 💬 1 📌 0
🌐 Explore & download:
• Check the new Local Ancestry tab in the gnomAD browser: gnomad.broadinstitute.org
• Full LAI VCFs & open-source pipeline: github.com/broadinstitu...
📰 Texas Children’s press release: www.texaschildrens.org/content/news...
🧪 Why it matters:
Traditional allele frequencies can mask ancestry-driven differences in admixed genomes. LAI can help clinicians & researchers:
• Refine variant interpretation
• Improve carrier & disease prevalence estimates
• Identify population-enriched risk variants
• ~81% of variants with LAI calls now have an updated gnomAD-wide max frequency (grpmax) — changing how some variants are classified.
• Variants once below the 5% ACMG BA1 benign threshold can now exceed it in ancestry-specific tracts → stronger evidence to reclassify VUS → Benign.
🔎 What’s new:
• For the first time, gnomAD includes haplotype-level allele frequencies for genetically inferred African/African American (n = 20,250) and Admixed American (n = 7,612) genomes.
• ~85% & ~78% of variants differ ≥2× across ancestry tracts, respectively.
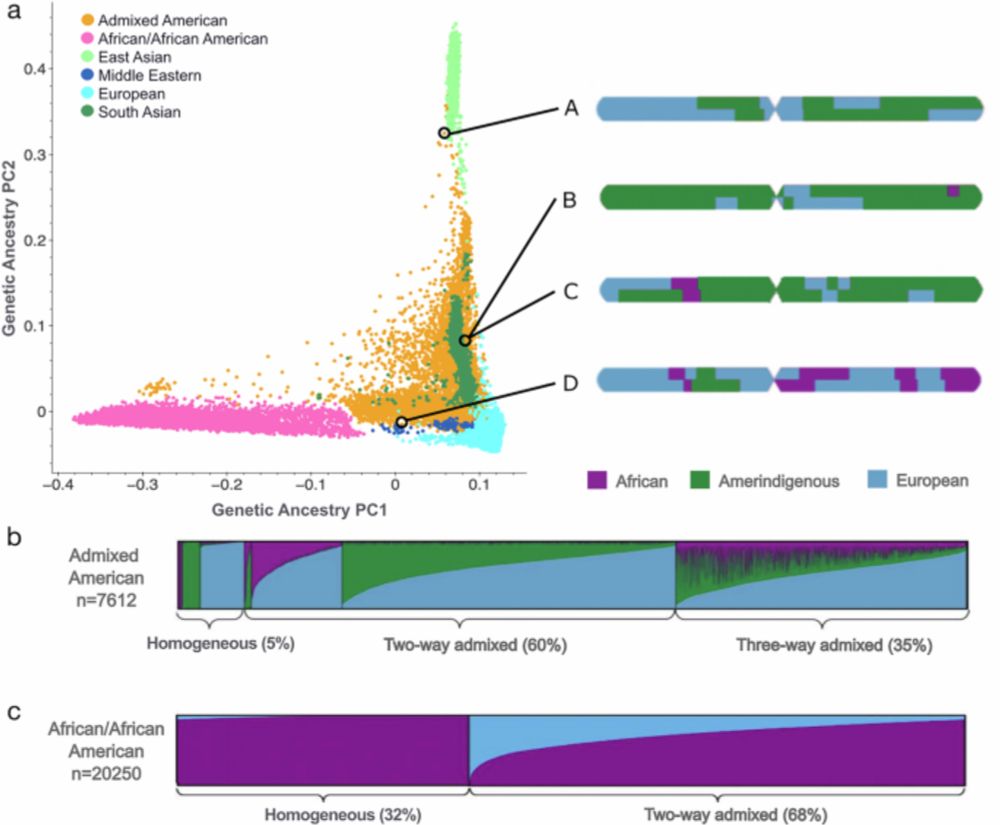
📃 We’re excited to share our latest work, now published in Nature Communications — a major update to the Genome Aggregation Database (gnomAD) that improves allele frequency resolution for two gnomAD-defined genetic ancestry groups using local ancestry inference (LAI).
06.10.2025 18:31 — 👍 29 🔁 8 💬 1 📌 1