See preprint for more on how genetic variation impacts the RBC proteome! Big thanks to a fantastic scientific village, including Gary Churchill @jacksonlab.bsky.social, Jim Zimring (UVA), and @dalessandrolab.bsky.social. Getting to work on this stuff sustains me in the current chaos we all face.
09.03.2025 19:34 — 👍 1 🔁 0 💬 0 📌 1

The hemoglobin beta locus is driven by a known genetic variant that induces an additional cysteine residue that is present in 5 of the founder strains. This locus regulated glutathione levels and drove a ptmQTL hotspot, highlighting the central role of hemoglobin in RBC metabolism.
09.03.2025 19:34 — 👍 0 🔁 0 💬 1 📌 0

Likely due to this, we observed a notable lack of cis-genetic regulation compared to other tissues previously studied in these mice. In its place, we observed strong trans pQTL hotspots, including at the hemoglobin alpha and beta. Many align with metabolite and lipid QTL (doi.org/10.1182/bloo...).
09.03.2025 19:34 — 👍 0 🔁 0 💬 1 📌 0

We profiled the red blood cell (RBC) proteome (including PTMs) and the impacts of RBC storage in a genetically diverse mouse population. The RBC is a fascinating and unique cell system, notably lacking nuclei and the ability to respond to stress via de novo protein synthesis.
09.03.2025 19:34 — 👍 1 🔁 0 💬 1 📌 0
Please check out heavily revised version of my manuscript comparing genetically diverse mouse populations (CC, CC-RIX, DO). I greatly expanded the simulations and now provide power curves!
x.com/grkeele/status…
02.01.2023 20:03 — 👍 0 🔁 0 💬 0 📌 0

GitHub - gkeele/musppr: Package to evaluate genetic analysis in mouse multiparental populations (CC, CC-RIX, and DO)
Package to evaluate genetic analysis in mouse multiparental populations (CC, CC-RIX, and DO) - gkeele/musppr
This manuscript was a fun side project for me. I hope it can provide some tangible examples and help others better utilize these powerful populations.
I also wrote an R package, musppr , that can be reused to tailor findings to others' experiments.
github.com/gkeele/musppr
29.08.2022 16:28 — 👍 0 🔁 0 💬 0 📌 0

Other interesting findings include how CC-RIX samples with replicates can better estimate additive heritability in the presence of F1-specific genetic effects. The CC can't tease these components apart, and when trying to estimate just the additive, returns the sum.
29.08.2022 16:28 — 👍 0 🔁 0 💬 1 📌 0
It's important to note that all populations have value for both heritability and QTL. The CC and CC-RIX had plenty of power for larger effect QTL (>=40%) -- good for eQTL, etc, and large DO samples provide meaningful, unbiased estimates of heritability.
29.08.2022 16:28 — 👍 0 🔁 0 💬 1 📌 0

In contrast, I expected the DO to be better at QTL mapping, particularly for small effect loci in polygenic traits. The extent of this was also surprising, with 174 DO mice being better power to detect a 10% QTL in a trait that is 90% heritable than 500 CC or CC-RIX mice.
29.08.2022 16:28 — 👍 0 🔁 0 💬 1 📌 0

My hunch was that experiments that included replicates (CC or CC-RIX) would more efficiently estimate heritability. The extent of this surprised me. For example, five replicates per 10 CC strains (50 mice total) had greater precision than 500 DO mice in some cases!
29.08.2022 16:28 — 👍 0 🔁 0 💬 1 📌 0

Interested in doing an genetic experiment with a cutting-edge mouse multiparental population (MPP), but don't know which best suits your needs (e.g., inbred vs outbred)?
Using simulations from real genetic data, I sought to provide some answers and guidelines.
x.com/biorxivpreprin…
29.08.2022 16:28 — 👍 0 🔁 0 💬 1 📌 1
Check out this cool work from Maddie evaluating how error can influence mediation inference. Something to consider when trying to causally relate -omic data with differing measurement error properties.
x.com/MS_Gastonguay/…
20.07.2022 18:27 — 👍 0 🔁 0 💬 0 📌 0
Really want to commend @tianzhang4921 for this work. Processing all the data, performing all the analyses, finding the message and the stories, and follow up. It's so much work. But that is at least balanced by how rewarding it is to share with our scientific communities now.
06.06.2022 15:34 — 👍 0 🔁 0 💬 0 📌 0
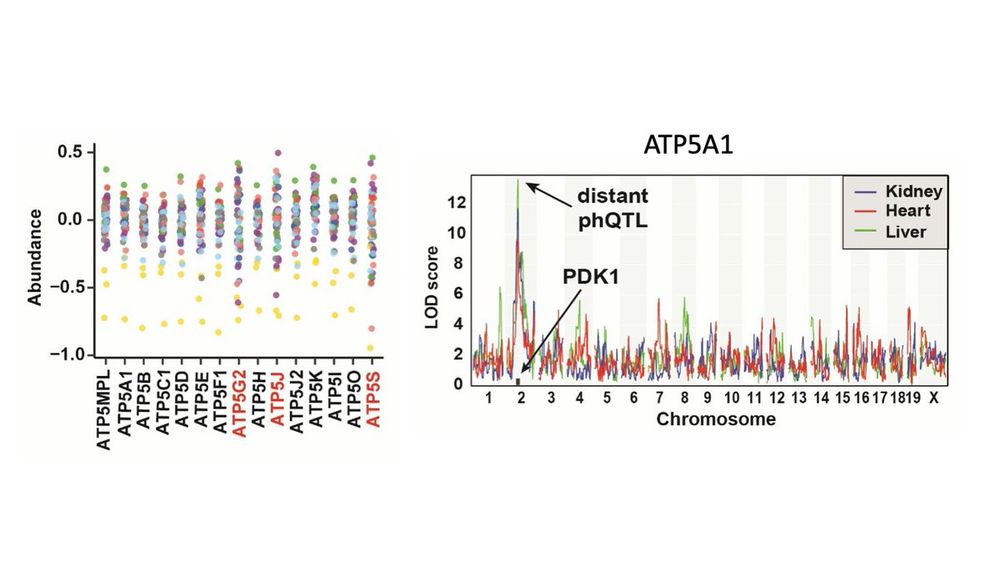
Other cool stories abound. Abundance of the ATP synthase in the heart appears to be stoichiometrically regulated through a low AJ allele at ATP5H. Independent of its protein abundance, phosphorylation of ATP5A1 is regulated by Pdk1.
06.06.2022 15:34 — 👍 0 🔁 0 💬 1 📌 0

The phos regulation through Pdk1 is particularly interesting because it is primarily driven by a low NZO allele. The NZO mouse is a very distinct polygenic model for obesity and diabetes, suggesting that unique regulation of phosphorylation could contribute to its phenotype.
06.06.2022 15:34 — 👍 0 🔁 0 💬 1 📌 0

We looked for genetic effects independent of parent protein by using a regression adjustment. This allowed us to identify distant phQTL that were mediated by plausible catalysts, such as kinases. For example, Pdk1 appears to phosphorylate 7 proteins across the tissues.
06.06.2022 15:34 — 👍 0 🔁 0 💬 1 📌 0

The abundance of a phosphopeptide was often determined by the abundance of its protein (which we refer to as the parent protein). Using a form of mediation analysis, we see that many phQTL essentially reflect an underlying pQTL, particularly when local.
06.06.2022 15:34 — 👍 0 🔁 0 💬 1 📌 0

This project builds from our previous work where we compared genetic effects on liver proteins among genetically diverse mouse populations.
Here, we integrate expression, proteins, and phosphopeptides from three tissues of 58 inbred CC mouse strains.
doi.org/10.1016/j.xgen…
06.06.2022 15:34 — 👍 0 🔁 0 💬 1 📌 0
Very grateful to have the opportunity to work with @tianzhang4921 dissecting how genetic variation affects protein phosphorylation. Our great team included @stevemunger, @MTFerris, @GygiLab, and Gary Churchill @jacksonlab. I'll share a few insights from the manuscript.
x.com/tianzhang4921/…
06.06.2022 15:34 — 👍 0 🔁 0 💬 1 📌 0
Paper alert! Final form of work with @wescrouse, from the labs of Gary Churchill @jacksonlab and @WilliamValdar. Includes fun examples of using bmediatR for genetic mediation analysis in genetically diverse mouse and human cell line data.
x.com/wescrouse/stat…
02.06.2022 17:46 — 👍 0 🔁 0 💬 0 📌 0
Aging B6 Proteomics - Home
I hope others find this study interesting and welcome feedback. It emphasizes the extent to which aging affects proteins post transcription.
Finally, these data represent a resource from the reference mouse strain. See RShiny app to explore yourself!
aging-b6-proteomics.jax.org
23.05.2022 17:09 — 👍 1 🔁 0 💬 0 📌 0

The effects of age and sex on proteins complexes can be, well, complex. We see changes in overall abundance due to age and sex, as well as changes to how tightly correlated complex members are. These differences can also be tissue-specific, as in the following ribosome complex.
23.05.2022 17:09 — 👍 0 🔁 0 💬 1 📌 0

Tissues also exhibit unique aging changes to functionally related proteins. In the spleen, for example, proteins increase with age that are related to the ER and protein trafficking. This also highlights co-regulatory signatures we see for protein complexes.
23.05.2022 17:09 — 👍 0 🔁 0 💬 1 📌 0

Across tissues, we see aging changes in immune protein levels. Immunoglobulins notably tend to increase with age. Even in tissues where the differences don't meet statistical significance, the direction of effects are consistent. We also see matching signal in immunoproteasomes.
23.05.2022 17:09 — 👍 0 🔁 0 💬 1 📌 0

The majority of age differences for proteins were not observed in their transcripts. This is in contrast to sex differences, which are often consistent.
We compared to transcript data from a separate study of a sister strain of B6. This matches our previous findings in DO mice.
23.05.2022 17:09 — 👍 0 🔁 0 💬 1 📌 0

We tested for differences in individual proteins' abundance due to age, sex, and age-by-sex. Tissues like kidney and liver had many proteins with sex differences, consistent with previous studies. Notably, many tissues had many proteins (approaching 500) with age differences.
23.05.2022 17:09 — 👍 0 🔁 0 💬 1 📌 0

We used multiplexed mass-spec proteomics to quantify protein abundance across 10 tissues from 20 C57BL/6J mice, representing a balanced factorial design in terms of sex and age (8 and 18 months) within a tissue. Age groups are roughly analogous to young adult and later midlife.
23.05.2022 17:09 — 👍 0 🔁 0 💬 1 📌 0
Excited to share this pre-print on how protein abundance changes with age across 10 tissues of B6 mice.
My awesome collaborators include @dschweppe1, Gary Churchill, Ron Korstanje @jacksonlab, and Steve Gygi @GygiLab.
I'll highlight some of the interesting findings below.
x.com/biorxivpreprin…
23.05.2022 17:09 — 👍 0 🔁 0 💬 1 📌 0
Personal account where sports, science, and my random thoughts intersect. Views are my own.
R, data, 🐕, 🍸, 🌈. He/him.
Bioinformatics engineer and scientist. Anything and everything related to improving DevX & research processes peak my interest
Geneticist, neuroscientist, behaviorist @Jackson Laboratory in Bar Harbor, Maine. www.kumarlab.org
Official account of the University of Nebraska–Lincoln, a top-tier research & Big Ten university. Home of the Huskers.
Genetics, bioinformatics, comp bio, statistics, data science, open source, open science!
Bren Professor of Computational Biology @Caltech.edu. Blog at http://liorpachter.wordpress.com. Posts represent my views, not my employer's. #methodsmatter
Est. 1929, JAX is a nonprofit scientific research institute specializing in genetics, genomics, mouse, and cellular models of disease. www.jax.org
Associate Dean for data science, and Professor of Medicine and Human Genetics at the University of Chicago.
Assistant Prof at D-BSSE, ETH Zurich, studying genetics of complex traits, with a focus on psychiatric disorders
www.nacailab.com
Bluesky newbie. Twitter exile. Previously Professor of Diabetes in Oxford, now heading human genetics at Genentech. Living the California dream. Views my own. #YNWA
Population geneticist, University of Chicago
Professor at Johns Hopkins Biomedical Engineering and CS, Director Malone Center for Engineering in Healthcare, Research Director (Strategy and Partnerships) Data Science and AI Institute
My lab at Stanford studies human population genetics and complex traits.
Senior Research Scientist @dfcidatascience @harvardchanschool
Cancer genomics, bioinformatics, chromatin & epigenetics, single-cell, plus occasional dog, food, and travel photos
AKA @jeremy_m_simon, @jeremy@genomic.social elsewhere
Professor, Dept of Pharmaceutical Sciences, Center for Drug Discovery, Northeastern University, Addiction Genetics, Neuroscience




















