15/ This of course wouldn't have been possible without the generous contribution of the patients and their families, for which we are ever thankful.
10.05.2025 22:56 — 👍 4 🔁 1 💬 0 📌 0
14/ Big thanks also to the institutions that provided tumor samples @dukepress.bsky.social, @mdanderson.bsky.social, Tokyo University Hospital, Pitié-Salpêtrière Hospital, St. Michael's Hospital, Seoul National University and NORLUX and funders NIH, NCI, Mark Foundation and Sontag Foundation.
10.05.2025 22:56 — 👍 5 🔁 2 💬 1 📌 0
13/ @weizmanninstitute.bsky.social, Mass General Brigham (Cancer Center, Pathology), @broadinstitute.org, @yalepress.bsky.social and University of Miami.
10.05.2025 22:56 — 👍 3 🔁 2 💬 1 📌 0
12/ Big thanks to the PIs that supervised the project @itaytirosh.bsky.social, @suvalab.bsky.social, @roelverhaak.bsky.social, Antonio Iavarone and Anna Lasorella, all collaborators and institutions involved in the CARE consortium >
10.05.2025 22:56 — 👍 4 🔁 2 💬 1 📌 0
11/ To sum-up, our multi-center study offers a high-resolution atlas of GBM recurrence dynamics, shaped by treatment response and TME context and underscores the tremendous heterogeneity of this devastating disease.
10.05.2025 22:56 — 👍 2 🔁 1 💬 1 📌 0
10/ Overall genetic divergence (SNVs + CNAs) correlated with transcriptional evolution, suggesting that GBM cell state shifts during recurrence are at least partially genomically driven.
10.05.2025 22:56 — 👍 3 🔁 1 💬 1 📌 0
9/ Small deletion phenotype (linked to radiotherapy) was associated with hypoxia-related states at recurrence, likely reflecting selection of radioresistant subpopulations. SBS21 (MMR-deficiency signature) also increased post-treatment in both CARE and GLASS cohorts.
10.05.2025 22:56 — 👍 3 🔁 1 💬 1 📌 0
8/ Interestingly, quantifying MGMT activity from the single-cell expression data outperformed promoter methylation as a prognostic marker in this cohort.
10.05.2025 22:56 — 👍 3 🔁 1 💬 1 📌 0
7/ However, in certain sub-groups, specific trajectories do tend to recur more often. First, MGMT-Low tumors (likely TMZ responders) tend to lose MES-like states whereas MGMT-High tumors (likely non-responders) tend to gain MES-like at recurrence.
10.05.2025 22:56 — 👍 3 🔁 1 💬 1 📌 0
6/ Despite global conservation, individual matched tumor pairs showed frequent divergence, with the majority switching at least one transcriptional layer. Overall, 72% of all possible transitions were observed across the cohort.
10.05.2025 22:56 — 👍 3 🔁 1 💬 1 📌 0
5/ Contrary to previous studies, we did not observe a significant enrichment of mesenchymal-like (MES-like) states at recurrence. Instead, recurrence trajectories were diverse and patient-specific, with no single state dominating across the cohort.
10.05.2025 22:56 — 👍 3 🔁 1 💬 1 📌 0
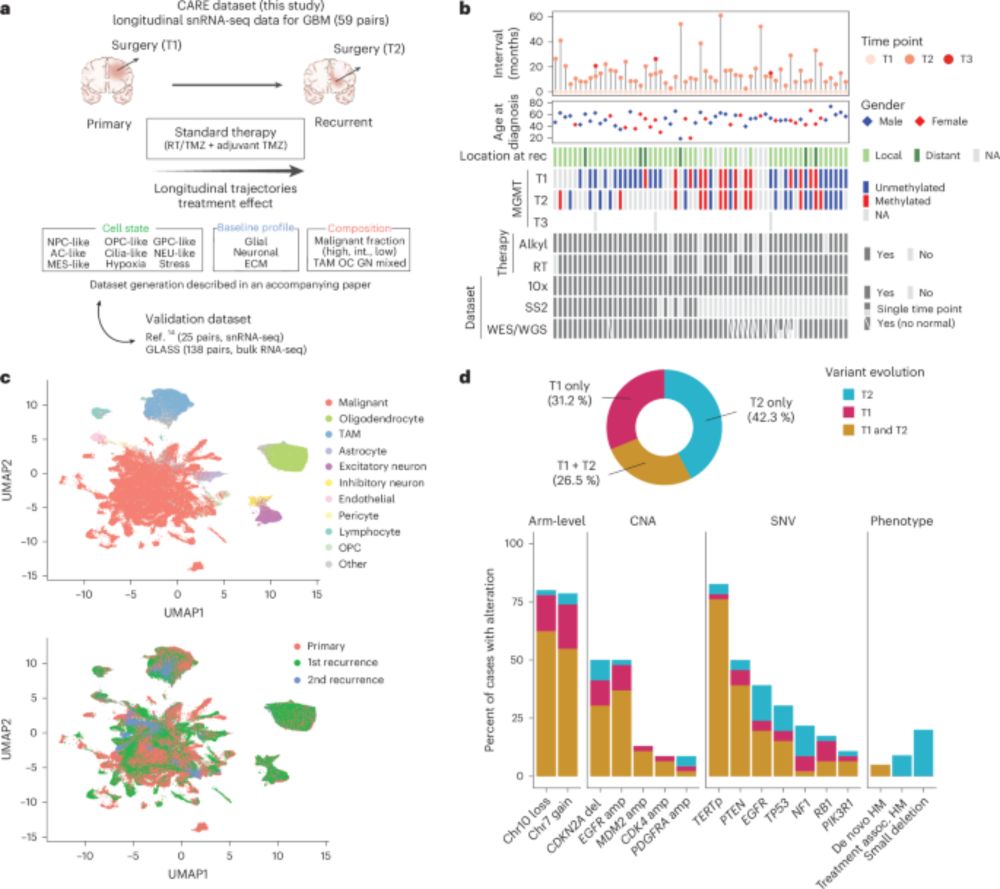
4/ And now for the results! Across the cohort the most consistent change at recurrence was reduced malignant cell fraction, with a reciprocal increase in glial and neuronal TME cells. This was observed in ~66% of patients, suggesting increased tumor integration into brain tissue.
10.05.2025 22:56 — 👍 4 🔁 2 💬 1 📌 0
3/ We first leveraged this large cohort to revisit the intra- and inter-tumor heterogeneity in GBM and characterized three multi-layered transcriptional ecosystems. Read more about this study in this great thread by @masashi-nomura.bsky.social →
bsky.app/profile/masa...
10.05.2025 22:56 — 👍 3 🔁 2 💬 1 📌 0
2/ In this study we profiled the longitudinal evolution of glioblastoma at single-cell resolution - altogether 121 treatment-naïve and exposed tumors from 59 patients, 430K nuclei, full clinical annotation and whole exome/genome sequencing.
10.05.2025 22:56 — 👍 3 🔁 1 💬 1 📌 0
PhD student | #Glioblastoma #scRNAseq | Tumor Heterogeneity
Cancer researcher at Hunter College of the City University of New York & Weill Cornell Medicine. Views are my own.
NY Times Besting Author of the Parkinson’s Plan, Medical Advisor Parkinson Foundation, Author 15 books http://pdplan.org Distinguished Professor and Director of the Fixel, Associate Editor JAMA Neurology, co-founder @DBSThinkTank
Physician scientist, Glioma single-cell genomics researcher, Neurosurgeon @Tokyo University, Postdoc in Suva lab @MGH/Broad
A passionate neuro-oncologist. Producer and Co-Host of “Neuro-Oncology: The Podcast” @TJUHospital @KimmelCancerCtr
Organizational principles of cancer | www.greenwald-lab.org at Lunenfeld-Tanenbaum Research Institute & Assistant Professor, Dept. of Molecular Genetics, University of Toronto | Mother of 4, lover of candy, art, & books
Computational biologist. Professor. Co-Director of Disease Mechanisms and Therapeutics Training Area
https://www.schlessingerlab.org/
@IcahnMountSinai. Opinions are mine. #DrugDesign #AI
Research Fellow at Dana-Farber Cancer Institute/Harvard Medical School @ Filbin Lab
Damon Runyon-St. Jude Fellow @DanaFarber.bsky.social @HarvardMed.bsky.social | Filbin Lab | Pediatric Neuro-Oncology | Runner | Tetris enthusiast.
Physician Scientist | @DanaFarber.bsky.social @Harvardmed.bsky.social | Postdoc in Neuro-Oncology Research @ Filbin Lab | Pediatrics Resident @UKEHamburg
Cutting-edge research, news, commentary, and visuals from the Science family of journals. https://www.science.org
Nature Portfolio’s high-quality products and services across the life, physical, chemical and applied sciences is dedicated to serving the scientific community.
Cell Press partners with scientists across all disciplines to publish and share work that will inspire future directions in research. #ScienceThatInspires
Clinical Computational Oncology @DanaFarber @harvardmed @broadinstitute | Chief - Population Sciences | Prev.: @Stanford @UCLA @UCSF | Discl.: http://bit.ly/2kF9C7B
Neurologist, Chair of Neurology
Brain Tumor Immunotherapy
@dkfz.bsky.social
@platten-lab.bsky.social
@uniheidelberg.bsky.social
Medical Faculty Mannheim
Founder Tcelltech
President @eanoassociation.bsky.social
Ambassador for the @erc.europa.eu
Research Scientist in Department of Neurosurgery Yale University studying glioma evolution
Professor of molecular systems biology at Karolinska Institutet, Sweden. On a mission to delete brain cancer using DNA therapy. Email: sten.linnarsson@ki.se
Scientist. Group leader at the Sanger Institute, Cambridge UK. Somatic mutation and selection in normal tissues, cancer and ageing.