A wee bit bumpier than Hamburg too. 😉
24.02.2026 14:21 — 👍 1 🔁 0 💬 1 📌 0A wee bit bumpier than Hamburg too. 😉
24.02.2026 14:21 — 👍 1 🔁 0 💬 1 📌 0
What a privilege to have this 5 minutes away from the office. Has spring sprung?
24.02.2026 13:46 — 👍 8 🔁 0 💬 1 📌 0Looks great!
23.02.2026 09:18 — 👍 1 🔁 0 💬 0 📌 0
Since last year, I've been making a RELION GUI that supports viewing, navigating and running jobs in a way completely compatible with the original RELION pipeline. Now it's working nicely for both SPA and tomography! Please try it out - github.com/hanjinliu/hi...
22.02.2026 20:10 — 👍 84 🔁 28 💬 4 📌 0If you're interested in how proteins of the innate immune response recognise and bind to pathogens (and/or how some microbes might avoid detection...), I invite you to read the paper now in press at the Journal of Biological Chemistry... www.jbc.org/article/S002... #crystallography
20.02.2026 17:47 — 👍 11 🔁 3 💬 0 📌 0Paper accepted! 🎉 More Thursday’s like this, please.
19.02.2026 20:17 — 👍 9 🔁 0 💬 0 📌 0
We absolutely love Krios days. Screening grids, collecting data will get old.
09.02.2026 16:26 — 👍 4 🔁 0 💬 0 📌 0And now out in Nature Comms (congrats Wyatt et al.): www.nature.com/articles/s41....
27.01.2026 15:27 — 👍 12 🔁 3 💬 0 📌 0Cryo-EM structure reveals how influenza A virus NEP binds the viral polymerase at a regulatory hotspot, coordinating RNA synthesis and nuclear export. Fantastic collaboration with @loiccarrique.bsky.social and Jon Grimes. www.science.org/doi/10.1126/...
24.01.2026 17:25 — 👍 54 🔁 27 💬 1 📌 0
Organelles do NOT have a single uniform pH.
And if you think they must, because “protons diffuse fast,” this paper is for you.
A thread on why that assumption is wrong; and what we found instead. 🧵 1/n
Experimental structures provide valuable hypotheses and accelerate but do not replace evolutionary and statistical analyses of protein sequences
15.12.2025 09:38 — 👍 38 🔁 9 💬 1 📌 1An integrated workflow for structural virology with a 100 keV electron microscope www.biorxiv.org/content/10.64898/2025.12.08.693081v1 #cryoEM
11.12.2025 09:16 — 👍 6 🔁 7 💬 0 📌 0Congrats Dan!
02.12.2025 16:32 — 👍 1 🔁 0 💬 0 📌 0As usual, there's a lot more cool stuff in this paper so naturally I encourage you to have a look! Thanks of course go to the entire team at EPFL, including the wonderful people at the DCI-Lausanne imaging centre who helped us collect something a lot of data.
22.11.2025 11:26 — 👍 1 🔁 0 💬 0 📌 0At 13.3 ms, we observe ErNaR in the O2 state. The chromophore has switched back to the all-trans configuration. We find the sidechain environment in the active center is starting to re-form. There also appears to be conformational dynamics taking place downstream along the proposed ion path.
22.11.2025 11:26 — 👍 0 🔁 0 💬 1 📌 0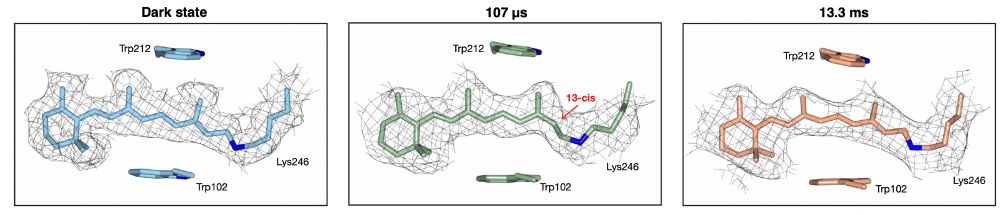
At 107 µs after photoactivation, we observe the K2 state. As we show, the chromophore has switched from the all-trans configuration present in the dark state to the 13-cis configuration. A switch that is accompanied by sidechain movements around the chromophore in the ErNaR active center.
22.11.2025 11:26 — 👍 0 🔁 0 💬 1 📌 0Samples of ErNaR were activated with a nanosecond laser pulse and subsequently jet vitrified at 107 µs and 13.3 ms, to probe the K2 and O2 states of the photo-cycle, respectively. We also collected a dark state, which was not photo-activated, to serve as a control in our experiments.
22.11.2025 11:26 — 👍 0 🔁 0 💬 1 📌 0
Next, to demonstrate that we could conduct time-resolved cryo-EM experiments, we turned to the bacterial rhodopsin ErNaR. Normally, ErNaR pumps sodium ions from the cytoplasm across the bacterial membrane to the ouside. Using our setup, we wanted to probe some of the earlier photocycle states.
22.11.2025 11:26 — 👍 1 🔁 0 💬 1 📌 0To demonstrate this setup, we first prepared a sample of mouse heavy chain apoferritin. After the usual data collection and processing, we obtained a 1.3 Å volume demonstrating that we can produce high-quality cryo-samples. The volume for this is shown higher up in this thread (with sidechains).
22.11.2025 11:26 — 👍 1 🔁 0 💬 1 📌 0You can see these "timing lasers" and the "time of arrival laser" (or as I prefer to call it, the bouncy laser) working together here! In this video, the jet would normally come from the right hand side whereas the activation laser would be fired from the left hand side.
22.11.2025 11:26 — 👍 0 🔁 0 💬 1 📌 0We also have a "time of arrival laser". Informally, we call this the "bouncy laser" because it continuously bounces off the back-side of a grid during an experiment. When the jet reaches the grid, this laser is deflected and this gives us the actual time of arrival of the jet.
22.11.2025 11:26 — 👍 0 🔁 0 💬 1 📌 0In other words, if we are interested in observing dynamics at 1 ms post-activation, we will fire the activation laser -1 ms before the predicted time of arrival of the cryogen jet. Simples!
22.11.2025 11:26 — 👍 0 🔁 0 💬 1 📌 0Using this predicted time of arrival, we then fire the activation laser as the appropriate delay, which activates protein dynamics either by releasing a photoactive compound (e.g. caged-ATP) or exciting a photoactive component within a protein itself (as we demonstrate in this paper).
22.11.2025 11:26 — 👍 0 🔁 0 💬 1 📌 0You can see these lasers in the first figure of this thread and can be divided into two. First, we have two "timing lasers", which detect the progress of the jet as it moves towards the grid. These allow us to calculate on-the-fly the predicted time of arrival of the jet at the grid.
22.11.2025 11:26 — 👍 0 🔁 0 💬 1 📌 0
We also took high-speed video frames of a similar jet and you can see these in the figure here. Now, a big part of our experiments is time. We want to accurately and precisely arrest protein dynamics at given timepoints to observe biological phenomena. To do this, we use a combination of lasers.
22.11.2025 11:26 — 👍 1 🔁 0 💬 1 📌 0Here you can see the the cryogen jet traveling towards the grid, ultimately striking the grid and, in doing so, arresting protein dynamics.
22.11.2025 11:26 — 👍 0 🔁 0 💬 1 📌 0To conduct a time-resolved experiment, a sample grid is placed in the activation subunit of the jetting assembly, and protein dynamics are initiated with a laser pulse. The sample is then vitrified with a liquid ethane (or ethane/propane) jet that is generated in the jetting subunit.
22.11.2025 11:26 — 👍 0 🔁 0 💬 1 📌 0
Experimental concept: protein dynamics are initiated with a short laser pulse. As they unfold, the sample is vitrified with a time-delayed jet of a liquid cryogen, trapping the proteins in their transient configurations.
22.11.2025 11:26 — 👍 1 🔁 0 💬 1 📌 0Happy to see our latest work online. Here we present a new method for microsecond time-resolved cryo-EM based on jet vitrification! Details in the 🧵 below... we hope you enjoy reading! (Hint, there's some fun videos...) #cryoEM #structuralbiology
22.11.2025 11:26 — 👍 26 🔁 6 💬 1 📌 0
Always a good feeling!
19.11.2025 17:41 — 👍 4 🔁 0 💬 0 📌 0