Correlative MS Imaging for cellular identification and analysis of in situ cryo-ET www.biorxiv.org/content/10.1101/2025.09.16.676641v1 #cryoEM
19.09.2025 09:16 — 👍 12 🔁 7 💬 1 📌 1
Improved cryo-EM reconstruction of sub-50 kDa complexes using 2D template matching www.biorxiv.org/content/10.1101/2025.09.11.675606v1 #cryoEM
17.09.2025 08:49 — 👍 20 🔁 11 💬 0 📌 1
This one is a bit of a departure from the usual and definitely a work in progress!
We found that by using ab initio reconstruction at very high res, in very small steps, we could crack some small structures that had eluded us - e.g. 39kDa iPKAc (EMPIAR-10252), below.
Read on for details... 1/x
13.09.2025 00:04 — 👍 259 🔁 81 💬 18 📌 12
PickET: An unsupervised method for localizing macromolecules in cryo-electron tomograms pubmed.ncbi.nlm.nih.gov/40894555/ #cryoEM
04.09.2025 05:08 — 👍 4 🔁 2 💬 0 📌 0

Now published @cp-cellreports.bsky.social! Antibody-mediated TGF-β1 activation for the treatment of diseases caused by deleterious T cell activity: www.cell.com/cell-reports...
Fantastic collaboration & massive effort from Sophie Lucas Lab @ de Duve Institute. #CryoEM with @savvideslab.bsky.social.
02.09.2025 11:25 — 👍 8 🔁 4 💬 2 📌 0

Upper - cryo-EM reconstruction of stomatin AQP1 complex. Lower - reconstruction of stomatin-UT-B complex.
Happy to share a new preprint from the Clarke & Vallese (@fravallese.bsky.social) labs reporting #cryoem structures of two RBC stomatin complexes – with AQP1 & UT-B - continuing the SPFH theme from our recent vault preprint! This was a fun one and has been cooking for a while - read on for more! ⬇️
30.08.2025 14:10 — 👍 119 🔁 30 💬 8 📌 7

EFFORTOCRACY
Exploring the concept of Effortocracy, a society where rewards reflect effort, contrasting it with traditional meritocracy based on achievements.
Lucky for you I've coined yet another term... now before you get too excited, it is about meritocracy. The first simul-post on both the website: nonzerosum.games/effortocracy... and the podcast: podcasts.apple.com/nz/podcast/n... Choose whichever is least painful. #Meritocracy #Society #Policy #DEI
30.08.2025 02:01 — 👍 46 🔁 11 💬 8 📌 2
Exciting to see our protein binder design pipeline BindCraft published in its final form in @Nature ! This has been an amazing collaborative effort with Lennart, Christian, @sokrypton.org, Bruno and many other amazing lab members and collaborators.
www.nature.com/articles/s41...
27.08.2025 16:14 — 👍 304 🔁 109 💬 14 📌 11

The small GTPase Ran defines nuclear pore complex asymmetry
Nuclear pore complexes (NPCs) bridge across the nuclear envelope and mediate nucleocytoplasmic exchange. They consist of hundreds of nucleoporin build…
My PhD paper is in press @cellpress.bsky.social and now online: www.sciencedirect.com/science/arti...!
Contents: weird #NPCs in yeast and flies, #CLEM, in situ #cryoET and more!
Made possible by the amazing @becklab.bsky.social team at @mpibp.bsky.social and collaborators! #TeamTomo
18.08.2025 16:26 — 👍 121 🔁 28 💬 6 📌 3
We’re looking for a Research Support Officer to join @matteoall.bsky.social’s group, working with @piotrkolata.bsky.social to investigate nuclear-cytoskeletal coupling in mammalian cells using #cryoEM, #cryoET & other biochemical approaches.
www.jobs.ac.uk/job/DOE317/r...
Closes 31 AUG
#ScienceJobs
11.08.2025 14:42 — 👍 7 🔁 3 💬 0 📌 0
We are excited to announce the first in-cell structure of a LINC complex within a native nuclear envelope at subnanometer resolution!! #In-cell cryo-ET + #atomistic MD simulations! @tomdendooven.bsky.social et al!!
www.biorxiv.org/content/10.1...
05.08.2025 12:41 — 👍 42 🔁 14 💬 2 📌 1
Check out our latest work now out on biorxiv www.biorxiv.org/content/10.1...
05.08.2025 11:27 — 👍 10 🔁 4 💬 1 📌 0
Congrats Kashish & everyone involved! Awesome stuff!
05.08.2025 11:58 — 👍 1 🔁 0 💬 0 📌 0

Attendees of the recent bacteriorhopsin symposium, photographed from above.
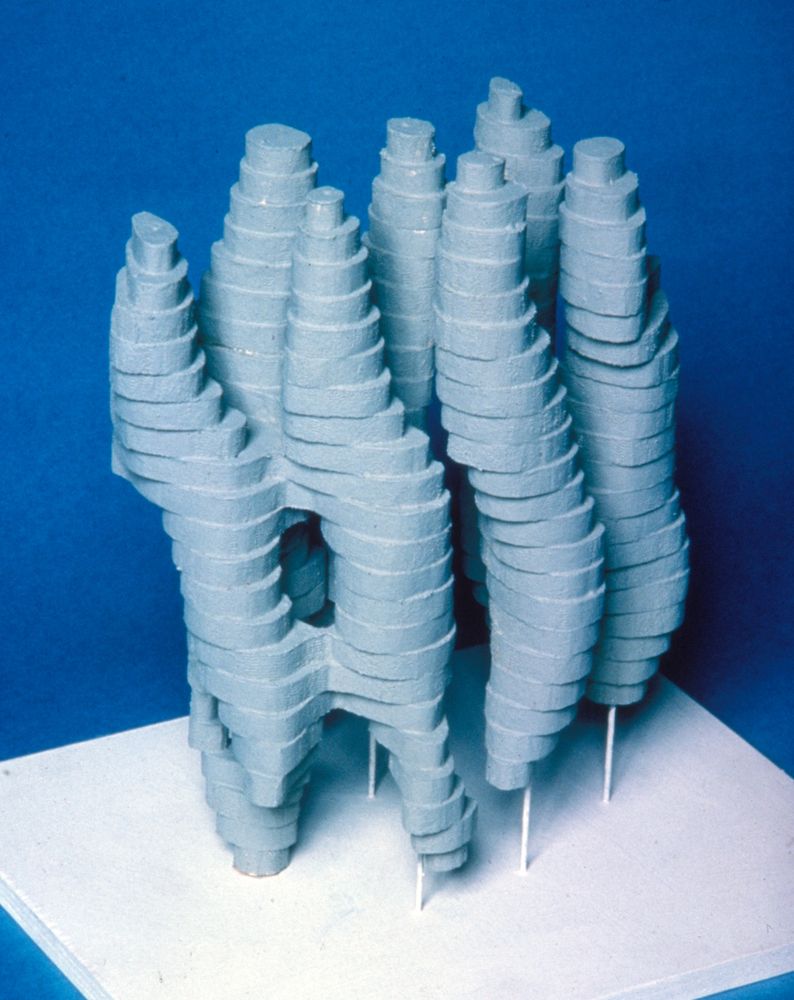
Balsawood model of bacteriorhodopsin

Black and white photograph of Nigel Unwin and Richard Henderson sitting outside the LMB, 1991

Symposium attendees applauding in the LMB's Max Perutz Lecture Theatre.
The LMB recently hosted a symposium to mark 50 years since Richard Henderson and Nigel Unwin published the structure of bacteriorhodopsin, the first 3D structure of a protein determined by #cryoEM.
Read about the event and find more photos here: www2.mrc-lmb.cam.ac.uk/the-structur...
#LMBNews 🧪🔬
04.08.2025 14:32 — 👍 57 🔁 10 💬 1 📌 2
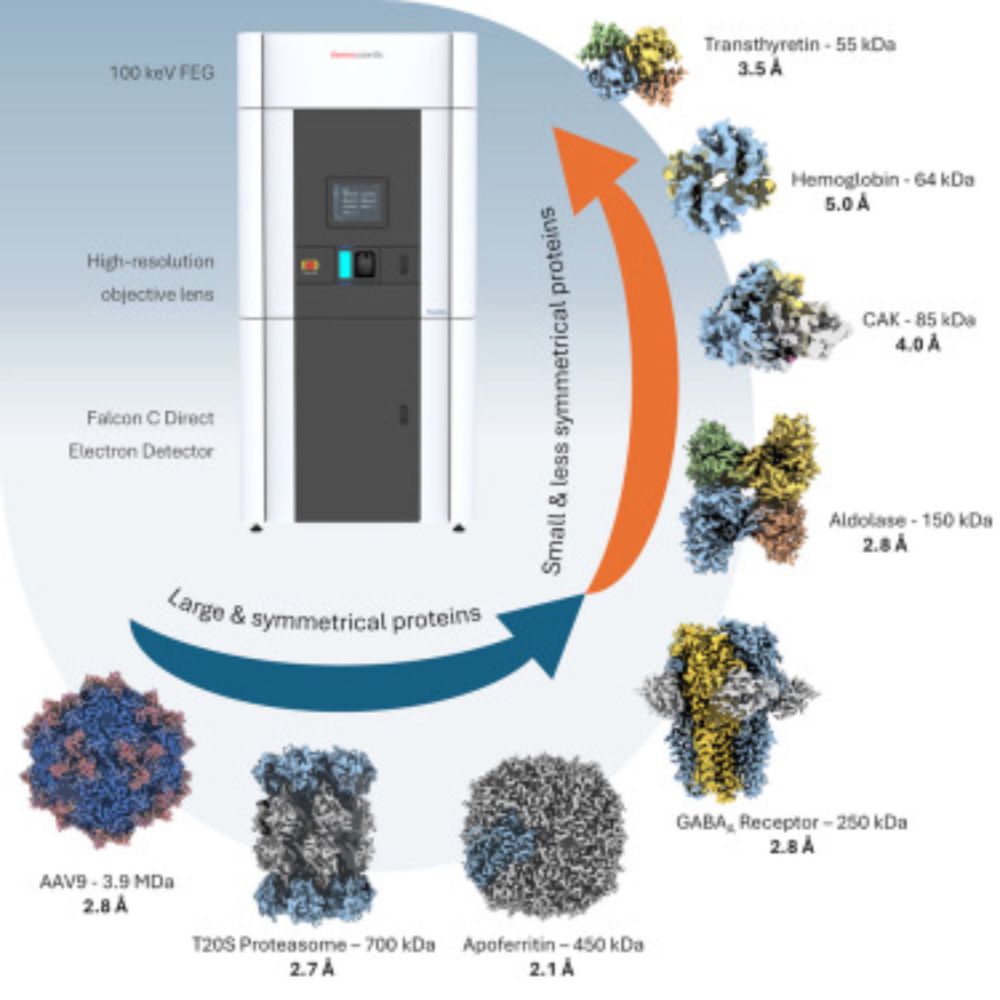
Sub-3 Å resolution protein structure determination by single-particle cryo-EM at 100 keV
Cryoelectron microscopy (cryo-EM) has transformed structural biology by providing high-resolution insights into biological macromolecules. We report s…
Now out in Structure: Paper on high-resolution #cryoEM using the 100 kV Tundra cryo-TEM by Dimple Karia, Adrian Koh, @abhaykot.bsky.social, @lingboyu.bsky.social, @landerlab.bsky.social and many others. Happy to have contributed to this effort. Check it out:
www.sciencedirect.com/science/arti...
30.07.2025 21:50 — 👍 80 🔁 37 💬 1 📌 0
Cool amyloid structures! Congrats Braja and the team!
26.07.2025 21:01 — 👍 1 🔁 0 💬 0 📌 0
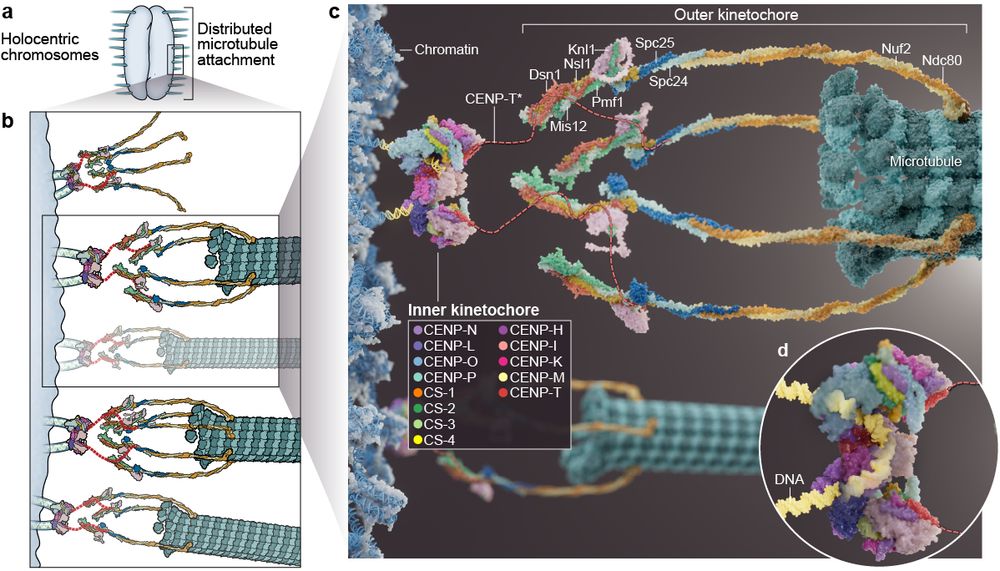
Happy to share our latest work on the structure and assembly of holocentric kinetochores! Huge thanks to Ines for a very fruitful collaboration, Claudio for all the support, and congratulations to Christine and all co-authors!
www.biorxiv.org/content/10.1...
15.07.2025 16:36 — 👍 71 🔁 24 💬 7 📌 6
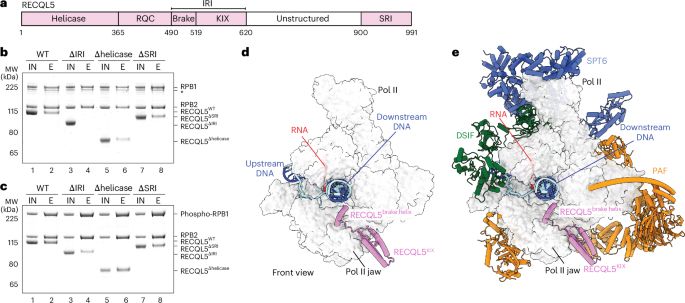
Excited to share our latest work! We found transcription factor SPT6 and phosphorylated Pol II CTD help recruit U1 snRNP to elongating Pol II, allowing efficient co-transcriptional splicing.
Glad to be featured in the Editors’ Highlights!
www.nature.com/articles/s41...
#splicing #cryoEM
10.07.2025 14:29 — 👍 69 🔁 21 💬 1 📌 0
Congratulations Jan & the team, very cool work!
10.07.2025 08:50 — 👍 0 🔁 0 💬 1 📌 0
A novel "bio-tag" for cryo-EM studies based on the small, electron-dense protein Csp1 pubmed.ncbi.nlm.nih.gov/40620219/ #cryoEM
09.07.2025 04:32 — 👍 14 🔁 7 💬 0 📌 0
New cryo-EM lore just dropped ft Niko and lab
#SmallStackHighRes #2DTM #cryoEM
....
For educational purposes only, copyright: 300 (film), Warner Brothers
07.07.2025 18:27 — 👍 9 🔁 2 💬 1 📌 0

Running Phenix and stuck?
Now you can ask the new Phenix chatbot anything from the 600-page tutorial about how to run any program.
Big thanks to Tom Willinger for making this happen!
Try it out: phenix-online.org/version_docs...
23.06.2025 05:14 — 👍 27 🔁 10 💬 1 📌 0
Our new preprint revealing a new mechanism of mitochondrial disease pathophysiology: the accumulation of toxic complex I intermediates bound to complex III
www.biorxiv.org/content/10.1...
19.06.2025 18:55 — 👍 9 🔁 3 💬 0 📌 0
Award-winning Science events for kids. Founded Ireland☘️Scientist & Mum, speaker United Nations 🇺🇳 Hands-on, STEM fun to kids Ireland 🇮🇪 UK 🇬🇧Canada 🇨🇦 Join us as Franchise Owners! Licences for International independent Schools junioreinsteinsscienceclub.com
Join a bleeding-heart liberal and compulsive speculator rambling about saving the world with win-win games at nonzerosum.games!
Co-Creating Ireland's Public Involvement in Open Research Roadmap
ENGAGED is building a national roadmap to shape public involvement in open research in Ireland. We believe that research can and does play an important role in tackling societal challenges.
CryoEM, membrane proteins and whatnot
Microscopy Manager @ University of Cambridge, Departments of Pathology & Biochemistry
Scientist and medical doctor. Gene regulation, DNA sequence models, transcription factor binding. Doing a PhD in computational biology at @molgen.mpg.de.
PhD student @MPI Science of Light
PhD candidate in Cellular Architecture group at IGBMC #structure_biology #biophysics
#cryo_ET #FIB_SEM #CLEM
Cellular Structural Biology Wellcome DPhil Student at The University of Oxford.
Currently working in Peijun Zhang’s lab at The Oxford Division of Structural Biology.
Passionate about biomedical animation and science communication: Batisio.co.uk
Ph.D Student @Schuller Lab @Uni_Marburg | Budding Structural Biologist | McGill University Alumnus | #Photosynthesis 🌱 #CryoEM❄🔬 #Biochemistry🧬 🇮🇳->🇨🇦->🇩🇪
Postdoc in the Carter Lab at the MRC-LMB. EMBO fellow. Alumnus of the Brouhard Lab.
Group leader & MRC CDA fellow at the Dunn School, Oxford. Cilia, motor proteins & intracellular organisation using cryo-EM/ET & cell biology. Run a co-lab with Anthony Roberts.
Studying cellular #stress responses, particularly #senescence and its impact on #immune response, #ageing and #cancer, #tumorigenesis at Cancer Research UK CI @cruk-ci.bsky.social, University of Cambridge @cam.ac.uk
Website: naritalab.com
Postdoc @Bharat group @MRC-LMB
Archaea, Eukaryotes, DNA replication & repair, Evolution enthusiast.
Biochemist, Structural biologist.
Postdoc #UniKiel
Unfocused wildlife 📸, Smasher of🏸. Opinions are my own
Taking care of Electron Microscopy @mpibp.bsky.social. Yay for #cryoEM and #cryoET ❄️🔬but frozen is not always better. Also, #viruses 🦠 rule.
cryoEM, structural biology, ribosomes
https://www.genzentrum.uni-muenchen.de/research-groups/beckmann/index.html
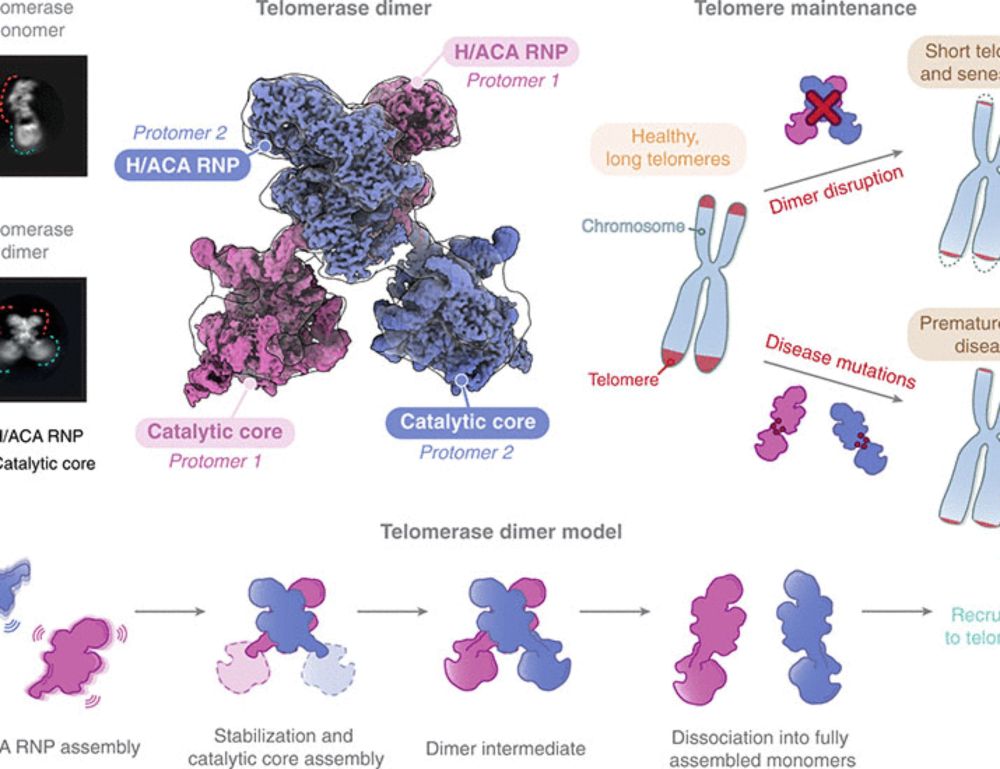
Postdoc at Masaryk University, Brno, Czechia | Advanced microscopy, Single-molecule biophysics, Telomeres, Genome maintenance.
Dad, Husband & Scientist | Immunology, Inflammation, Chemokines & Atypical ChemoKine Receptors (ACKRs) | Research & Meta-Research | University of Munich (LMU)
Molecular Inflammation Lab
https://duchene.ipek-research.com