🧵Excited to share our new preprint introducing iHOTT - an autologous tumor-immune co-culture model that captures patient-specific responses in #Glioblastoma
💥Now on
@biorxivpreprint
: biorxiv.org/content/10.1...
Led by Dr. Shivani Baisiwala, Neurosurgery Resident in the lab
24.06.2025 02:24 — 👍 6 🔁 4 💬 1 📌 0
Honored to join this amazing community 🎉
02.06.2025 19:34 — 👍 2 🔁 0 💬 1 📌 0
Excited to present our new preprint led by @claudianguyen95 uncovering how thalamic input shapes human cortical development! We discover that thalamic axons promote the generation of upper layer cortical neurons through NRXN1-mediated contacts with outer radial glia. www.biorxiv.org/content/10.1...
06.05.2025 16:50 — 👍 5 🔁 2 💬 1 📌 0
Very grateful to my mentor Aparna @bhadurilab.bsky.social , co-authors @elisafazzari @claudianguyen95 @RyanKan20 @yoojuyoun.bsky.social
@amartija.bsky.social Daria Azizad, Brittney Wick and Maximilian Haeussler!
02.05.2025 00:39 — 👍 3 🔁 0 💬 1 📌 0
We’re excited to see how our tools can continue to reveal cell fate mechanisms from molecular maps of the human brain. 🧠🗺️✨
And to open this approach to other biological problems, we’ve put our scripts to make your own meta-atlas and modules at github.com/BhaduriLab/d.... (13/13)
02.05.2025 00:39 — 👍 3 🔁 0 💬 1 📌 0
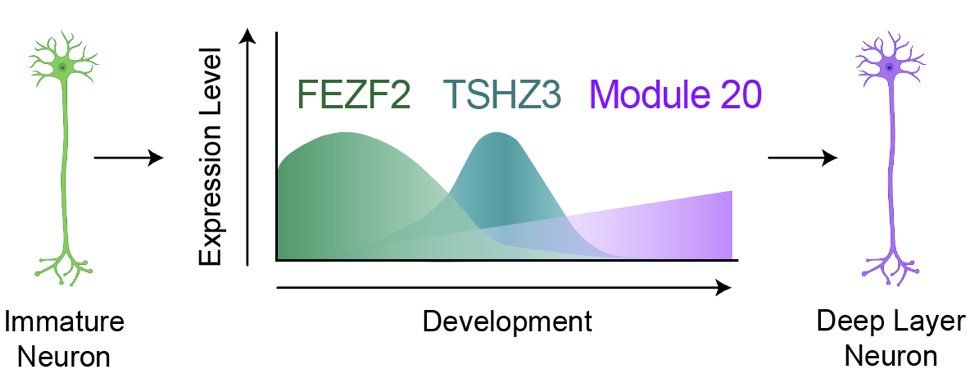
Even a subtle KD of these transcription factors produced a modest decrease in module 20 that cascaded into substantial composition differences. (12/13)
02.05.2025 00:39 — 👍 2 🔁 0 💬 1 📌 0
With chimeroids, we knocked down FEZF2 & TSZH3 across multiple genetic backgrounds. Consistently, both FEZF2 & TSHZ3 were required to make deep layer neurons – w/ FEZF2 acting at the level of gene expression and TSHZ3 acting at the chromatin level. (11/13)
02.05.2025 00:39 — 👍 2 🔁 0 💬 1 📌 0
Interestingly, TSZH3 and the rest of module 20 was less specific to FEZF2 subtype neurons in the developing and adult mouse brain. So we tested our model in human stem cell derived cortical chimeroid models developed by @bolanosanton.bsky.social and @irenefaravelli.bsky.social @ArlottaLab. (10/13)
02.05.2025 00:39 — 👍 2 🔁 0 💬 1 📌 0
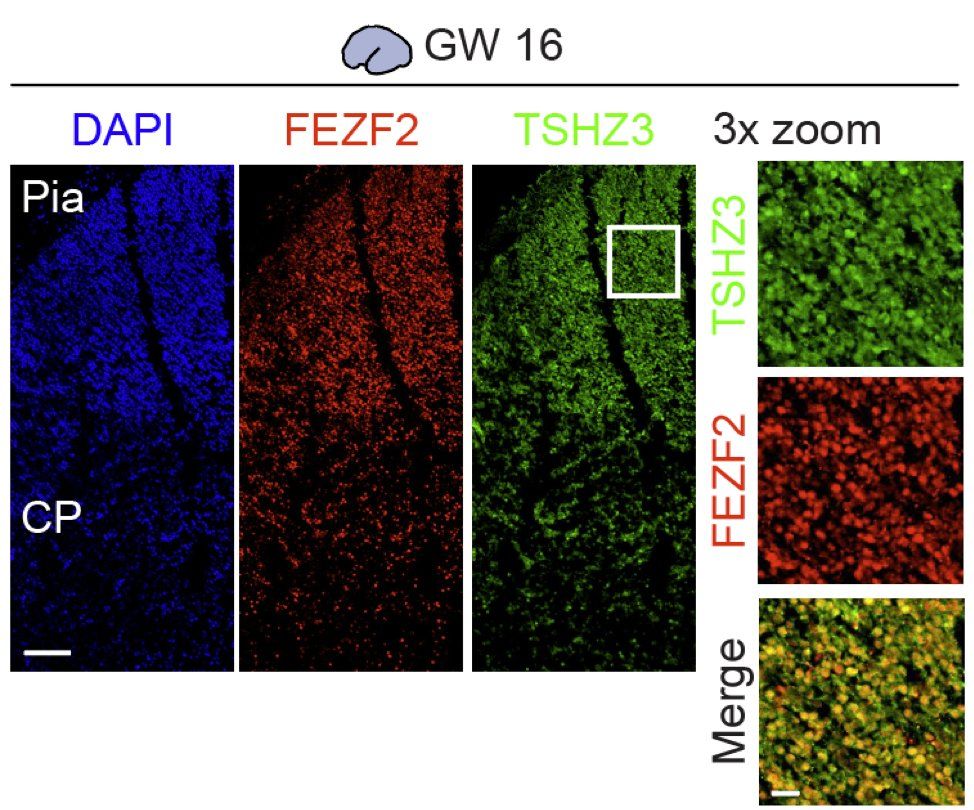
We validated that at GW16, FEZF2 and TSHZ3 co-express in deep layers of the human cortex, and by GW20, levels of FEZF2 decrease while TSHZ3 expression stays on in these deep layers. (9/13)
02.05.2025 00:39 — 👍 2 🔁 0 💬 1 📌 0
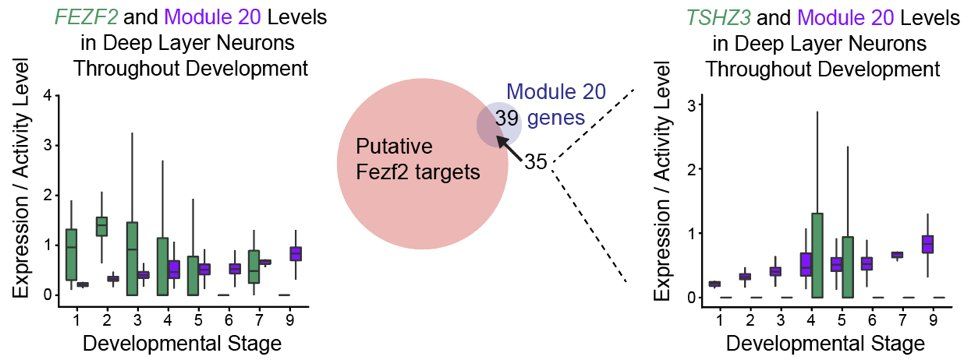
However, FEZF2 expression peaks before module 20 activation during development, and half of module 20 genes are candidate targets of FEZF2 (thanks to data from @LodatoS_Lab @ArlottaLab). This included one transcription factor, TSHZ3, linked to autism spectrum disorders. (8/13)
02.05.2025 00:39 — 👍 2 🔁 0 💬 1 📌 0
Most notable is module 20, which we determined drives the specification of deep layer neuronal subtypes found in the adult. Module 20 is enriched in human adult FEZF2 subtypes – but it doesn’t actually contain FEZF2. (7/13)
02.05.2025 00:39 — 👍 2 🔁 0 💬 1 📌 0
We found modules that may explain how neuronal/glial fates are initiated and refined into cell types found in the adult human cortex, with the spatiotemporal expression patterns predicted for some of these modules validating in primary tissue. (6/13)
02.05.2025 00:39 — 👍 2 🔁 0 💬 1 📌 0
We first annotated these modules for their cell type/biological function, then scored how each of these modules act in other datasets – ie the adult human brain, the developing mouse, and the adult mouse. @alleninstitute.bsky.social @danielajdibella.bsky.social (5/13)
02.05.2025 00:39 — 👍 3 🔁 0 💬 2 📌 0
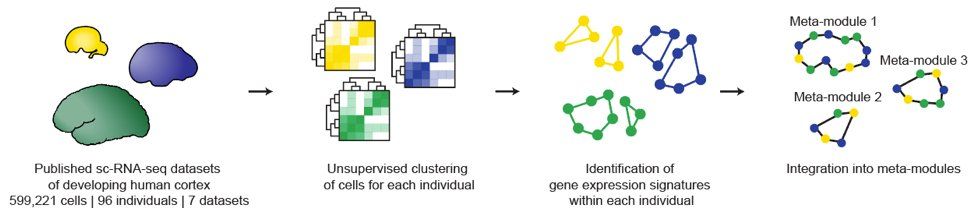
We used these atlases to find gene networks that shape cell types.
Step 1: Define networks w/in each indiv’d in our dataset w/ hierarchical clustering.
Step 2: Correlation analysis btwn indiv’d to id which networks co-expressed across the entire meta-atlas – these are our meta-modules. (4/13)
02.05.2025 00:39 — 👍 3 🔁 0 💬 1 📌 0
So we merged recently published datasets into two meta-atlases: one for the developing human cortex (7 datasets, 0.6M cells) & one for the adult human cortex (16 datasets, 2.6M cells) – both on the UCSC Cell Browser now:
dev-ctx-meta-atlas.cells.ucsc.edu
adult-ctx-meta-atlas.cells.ucsc.edu
(3/13)
02.05.2025 00:39 — 👍 1 🔁 0 💬 1 📌 0
Plenty of work has cataloged the cell types found in both the developing and adult human brain. But it’s hard to generate an atlas with enough scope and depth to clarify how the cell types present in development turn into those found in adulthood (2/13).
02.05.2025 00:39 — 👍 2 🔁 0 💬 1 📌 0
It’s out! The first paper from my postdoc – and first from the @bhadurilab.bsky.social – is now live @natneuro.nature.com . 🧠✨
Using a new meta-atlas generation strategy, we identified functional gene networks that more fully explain how cell types are formed in the human cortex. (1/13)
02.05.2025 00:39 — 👍 26 🔁 10 💬 3 📌 2
Postdoc in the Calipari lab | Social Media for Stories of WiN | Leading Edge Fellow | Topo Chico Fanatic
Neuroscientist, Sestan Lab @yaleneuro.bsky.social, Brain Development and Evolution, Human-Specific Features, #Cerebellum, Pluripotent Stem Cells, #iPSC, Individual Variation
UCLA Associate Professor, PhD Researcher of brains 🧠 (development, stem cells, neuroinflammation, autism, sensory processing, brain injury & repair)
Teacher of Neuroanatomy, Neurophilosophy (consciousness, cognitive science), & Stem Cell Biology
Developmental Neuroscientist and Stem Cell Biologist. My group and I study the mechanisms controlling neural development, circuit assembly, and disease.
Postdoc @ Puglisi Lab at Stanford University
HIV | RNA | ribosome | antibiotics | structural biology 🔬💊👩🏻🔬.
Weizmann Institute of Science Yonath Lab alum
Leading Edge Fellow 2025
BME UNITE (Underrepresented Needs In Technology & Engineering) is a national group of biomedical engineering faculty who are interested in educating themselves, improving representation, and combating racism in STEM. https://bmeunite.org/
Tiampo Family Associate Professor at Tufts University, cancer and epilepsy researcher, fierce advocate for diversity and inclusion, mom of a daughter with the SCN8A epilepsy
Mitochondrial biology and cancer ••• Postdoctoral fellow at
NYU Langone Grossman School of Medicine. Emerald Foundation Inc. Fellow. Leading Edge Fellow 2024 |Philips lab| +she/her+
Neuroscience, meninges, blood-brain barrier, cake (or cookies)
http://siegenthalerlabcu.weebly.com/
neuroscience postdoc in NYC (Froemke lab @ NYUMed). audition👂2P imaging 🔬and developmental systems neuroscience 👶. Stories of WiN co-founder/co-director, marathon runner, foodie, IPA lover, and overly enthusiastic dog mom
Computational Scientist at Octant Bio
Understanding life. Advancing health.
Epigenetics & Genomics
Harvard University & Broad Institute
https://www.macfound.org/fellows/class-of-2023/jason-d-buenrostro
buenrostrolab.com
Building neurotech to study how genes and physiology impact brain function 🧬 🧠 Associate Professor, Scripps Research | Freeman Hrabowski Scholar, HHMI
Developmental neurobiologist and human organoids aficionado. Associate Prof. @USCStemCell. Director @USC CIRM ASCEND Shared Resource Lab. Mama^2. Immigrant 🇮🇹.
https://quadratolab.usc.edu/
https://ascend.usc.edu/
Assistant Professor at at Stanford Biology & Stem Cell Institute. Studying human brain development through genomics and proteomics. Website: www.liwanglab.org
Assistant Professor at Emory University, Department of Human Genetics. Using organoids to study spinal cord development and degeneration
Cell Press partners with scientists across all disciplines to publish and share work that will inspire future directions in research. #ScienceThatInspires



