Our paper demonstrating that within-species warfare interactions are ecologically important on human skin is now published in Nature Micro! www.nature.com/articles/s41...
30.06.2025 12:26 — 👍 209 🔁 97 💬 9 📌 3

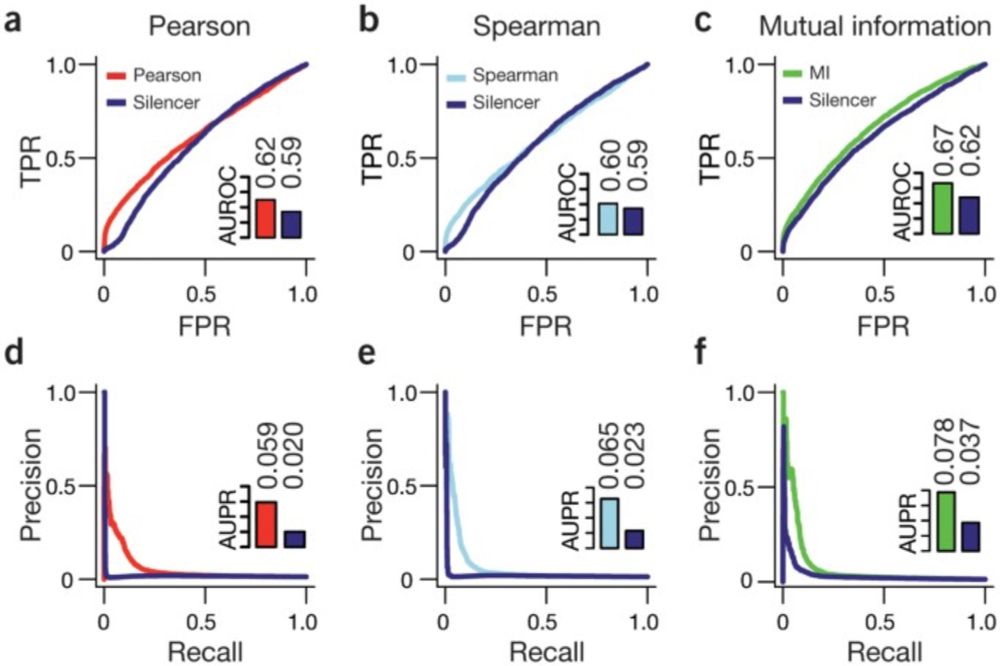
The paper has the incorrect dynamics for gLV
12.07.2025 17:21 — 👍 0 🔁 0 💬 0 📌 0
The mbtransfer paper also has data leakage issues. In many of the results they apply DESeq2 Normalization in a pre-processing step (for all samples in a bulk process) and include samples which are later used as forecasting test samples when doing so.
12.07.2025 17:21 — 👍 1 🔁 0 💬 1 📌 0

When MDSINE2 was supplied actual read abundances the authors note “[w]hen the data were not asinh transformed, the mbtransfer model performed worse than either MDSINE2 or fido”
12.07.2025 17:21 — 👍 0 🔁 0 💬 1 📌 0
The apparent poor performance of MDSINE2 in the main text of the mbtransfer paper is entirely from misuse of the model and supplying it a table of "read" abundance that are not actually read abundances.
12.07.2025 17:21 — 👍 0 🔁 0 💬 1 📌 0
The nonlinearly transformed values also mess up how the latent dynamics (gLV modeling CFU/g in our case) are interpreting the taxa abundances and how they are changing over time.
12.07.2025 17:21 — 👍 0 🔁 0 💬 1 📌 0
MDSINE2 has a Negative Binomial noise model for reads. Seeing transformed "reads" with values between 1 and 10 tells the model that the noise is through the roof for all taxa!
12.07.2025 17:21 — 👍 1 🔁 0 💬 1 📌 0
Our paper is now open access as well.
29.05.2025 16:23 — 👍 4 🔁 1 💬 0 📌 0
Call for papers on the clinical microbiome - Nature Microbiology
Nature Microbiology has launched a joint collection on the clinical microbiome with Nature Communications, Nature Medicine and Communications Medicine.
This month's editorial is an invitation to submit papers on clinical and translational aspect of the microbiome.
This collection is a joint effort with @naturemedicine.bsky.social @natcomms.nature.com and @commsbio.nature.com
#MicroSky #MicrobiomeSky 🦠⚕️🧪⚕️
www.nature.com/articles/s41...
06.05.2025 19:37 — 👍 25 🔁 18 💬 3 📌 1
reviewer 3 or 2?
06.05.2025 22:51 — 👍 1 🔁 0 💬 0 📌 0

It is a great pleasure to listen to @seppekuehnlab.bsky.social
His talk is about ´Learning microbiome design principles from natural variation ´.
Below, the #liveSketch painting during the seminar and given at the end!
#ArtAndScience #MediationScientifique
05.05.2025 10:30 — 👍 5 🔁 3 💬 2 📌 0
Openings
Positions available in the Gibson Lab
Several Postdoctoral Fellow openings. All fellows have triple appointment at HMS,BWH, and Broad
- Biological sequence models (theory, design, and application)
- Learning single cell dynamics
- Bacteriotherapy design, “bugs-as-drugs” (using control theory principles)
gibsonlab.io/openings/
17.04.2025 14:02 — 👍 2 🔁 1 💬 0 📌 0
We are still accepting registrations for this event! Please come join us and hear about cutting edge microbiome research.
17.03.2025 20:05 — 👍 8 🔁 3 💬 0 📌 0
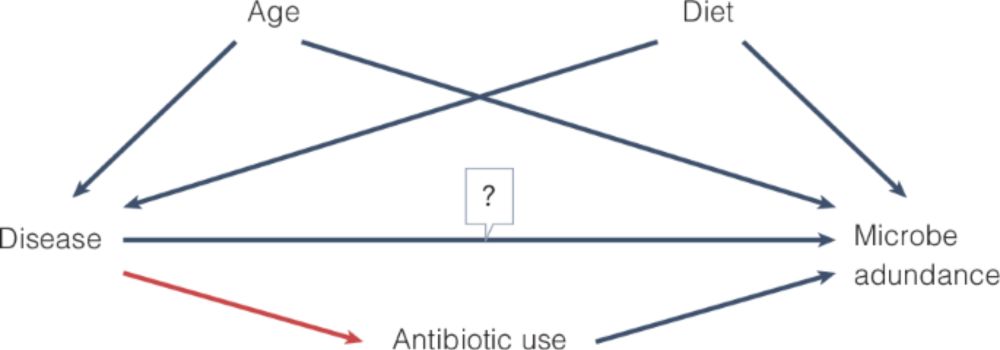
Planning and describing a microbiome data analysis - Nature Microbiology
We provide guidance on the planning, execution and description of statistical analyses in microbiome studies.
A must-read for any microbiome researcher 🦠📐and a new addition to our #bestpractices series
Planning and describing a microbiome data analysis by @amydwillis.bsky.social and @davidandacat.bsky.social
www.nature.com/articles/s41...
21.02.2025 14:48 — 👍 58 🔁 26 💬 1 📌 1
Paper just accepted
Gibson et al. "On the stability of gradient descent with second order dynamics for time-varying cost functions" Transactions Machine Learning Research (2025)
openreview.net/forum?id=Hlz...
07.02.2025 16:35 — 👍 2 🔁 1 💬 0 📌 0
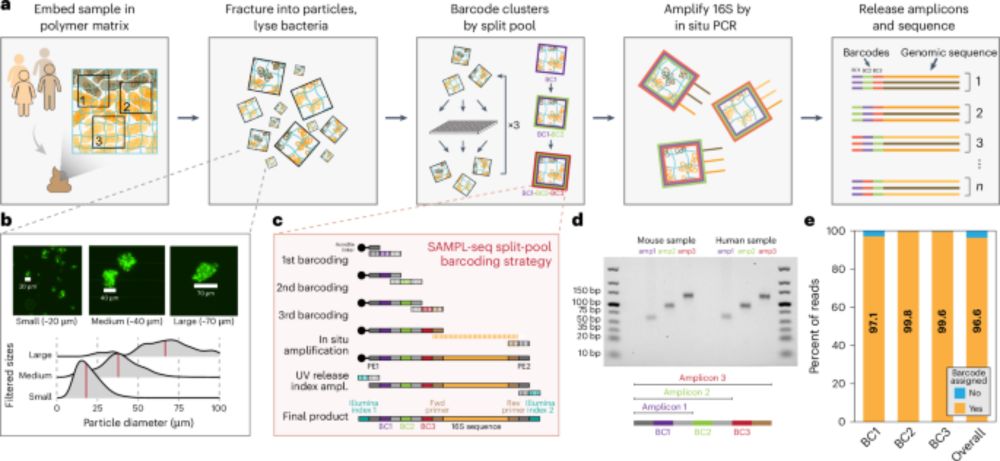
SAMPL-seq reveals micron-scale spatial hubs in the human gut microbiome - Nature Microbiology
Split-And-pool Metagenomic Plot-sampling sequencing (SAMPL-seq) can be applied to complex microbial communities to reveal spatial co-localization of microbes at the micron scale.
Happy to share our new paper in @naturemicrobiol.bsky.social on mapping spatial relationships of the human gut microbiome. We identified distinct spatial hubs between gut bacteria that reflect sub-community assemblies at the micron-scale. Led by Miles Richardson & co.
www.nature.com/articles/s41...
03.02.2025 14:46 — 👍 89 🔁 36 💬 0 📌 0
Are regret based adaptive control strategies deployable?
arxiv.org/abs/2501.04572
09.01.2025 14:38 — 👍 0 🔁 0 💬 0 📌 0
It’s easy to think this is about politics, but Ran’s actually just subtweeting my working style
08.12.2024 18:16 — 👍 13 🔁 2 💬 1 📌 0
As further evidence you should send them the @abrahamgihawi.bsky.social paper that invalidated and lead to the retraction of the Nature paper about a cancer microbiome.
03.12.2024 13:23 — 👍 5 🔁 0 💬 0 📌 0
at this point though we are speculating because there doesn't seem to be enough information in the paper or code for us to really see how the inference was actually performed
02.12.2024 20:15 — 👍 0 🔁 0 💬 0 📌 0
you can have leave one-out testing that respects the test train split... you just perform n rounds of train on (n-1) test on 1
02.12.2024 20:11 — 👍 0 🔁 0 💬 1 📌 0
This kind of mistake happens way too often
02.12.2024 19:58 — 👍 1 🔁 0 💬 0 📌 0
Postdoc at Dana-Farber and Harvard Med with Heng Li (@lh3lh3.bsky.social). Prev: UBC / UofT.
I like thinking about computational biological sequence analysis and its applications to metagenomics.
https://jim-shaw-bluenote.github.io
Postdoc at UCL with James Reading. Previously at EMBL working with Wolfgang Huber. Biostats, R, cancer immunology
HFSP Postdoctoral Fellow at MIT | Mobile genetic elements and microbial communities
Bioinformatics Group Leader - Department of Microbiome Science www.leylab.com - Max-Planck Institute for Biology, Tübingen
Big data, tiny microbes, endless questions. Let’s dive in 🌀
Postdoctoral Associate at the Xavier Lab at the @broadinstitute.org
All things host–microbe–microbe interactions.
Systems Biologist l Microbiome Research
Love everything host-microbe
Postdoctoral researcher at the University of Michigan, Ann Arbor
Instrumentation
Laboratoire Gulliver
Associate Professor
DFCI & HMS
Assistant professor @ Sloan Kettering Institute | Postdoc @ the Marraffini Lab Rockefeller University | PhD @ the Schuldiner Lab Weizmann Institute | microbial defense systems
Scientist, immigrant, mom (she/her)
We leverage tools from machine learning and control theory to advance our understanding of biological systems. (Harvard Medical School | Brigham and Women's Hospital)
PI: @travis.gibsonlab.io
Website: gibsonlab.io
A leading life science journal that champions high-impact research across all disciplines—from molecules to ecosystems. We offer innovative formats and collaborative editorial support to ensure your work achieves its full scientific impact.
Microbiome, metagenomics, ML, and reproductive health. All views are mine. So are all your base
UW biology prof.
I study how information flows in biology, science, and society.
Book: *Calling Bullshit*, http://tinyurl.com/fdcuvd7b
LLM course: https://thebullshitmachines.com
Corvids: https://tinyurl.com/mr2n5ymk
I don't like fascists.
he/him
Mathematician at UCLA. My primary social media account is https://mathstodon.xyz/@tao . I also have a blog at https://terrytao.wordpress.com/ and a home page at https://www.math.ucla.edu/~tao/
Computational and statistical genetics. Excited about all things science. Professor of genetics at UPenn. Views my own.
https://www.med.upenn.edu/bogdan-group/
Machine learning and biology. Research Scientist at Google DeepMind. adamgayoso.github.io. Views are my own.
Associate Professor at Princeton
Machine Learning Researcher
MRC Career Development Award fellow at the University of Cambridge | Group Leader of the Microbiome Function and Diversity lab (https://microfundiv-lab.github.io) | Bioinformatics, Machine Learning, Metagenomics, Functional modelling, Human microbiome