This looks fascinating, congratulations!
14.07.2025 08:13 — 👍 0 🔁 0 💬 0 📌 0Johannes Pilic
@johannes-pilic.bsky.social
Postdoc at ETH Zurich in the Kleele lab. I'm interested in mitochondria, microscopy and systems biology.
@johannes-pilic.bsky.social
Postdoc at ETH Zurich in the Kleele lab. I'm interested in mitochondria, microscopy and systems biology.
This looks fascinating, congratulations!
14.07.2025 08:13 — 👍 0 🔁 0 💬 0 📌 0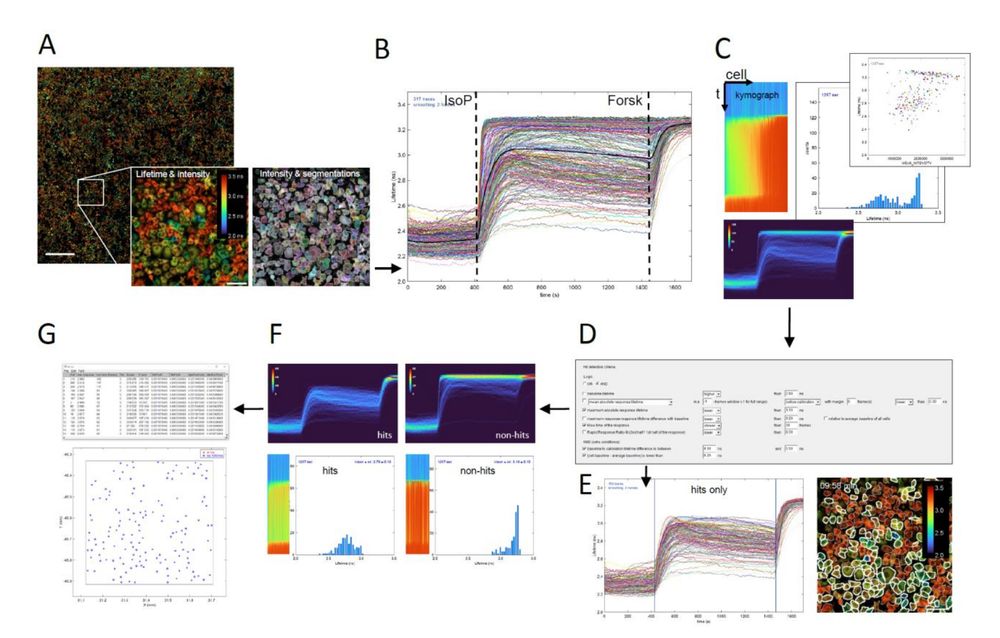
Fig. 4. Time-lapse FLIM analysis and hit detection Fiji macro. (A) Stitched multi-tile intensity image overlay with the fluorescence lifetime image. The lifetime image can be either average photon arrival times (‘Fast FLIM’) or calculated average lifetimes from two lifetime components. Ratiometric images can also be used. Scale bar: 1000 μm. Inset: Left – Zoom-in of lifetime image of cells after stimulation with isoproterenol; Right – Zoom-in showing cell segmentation as outlines. Scale bar: 100 μm. (B) Lifetime time traces of all 298 segmented cells in a single tile of Cos7 cells (a mix of ADRB2 KO and WT cells). Cells are stimulated with 40 nM isoproterenol (IsoP), followed by a calibration with forskolin (Forsk). Stimulation and calibration time points may be automatically calculated from the average of all traces (thick black line) or entered manually. (C) The script generates several informative visualizations. Shown are a ‘kymograph’ representation of fluorescence lifetime vs time in all cells, sorted on response magnitude (top- left); a normalized data density graph (2D histogram of lifetime vs time, with cell counts in false color), convenient when analysing thousands of cells (bottom); and (optionally animated) histograms of the fluorescence lifetimes over time (middle) and scatterplots of lifetime vs secondary parameters like cell intensity, cell area or intensity of an additional fluorescence channel of choice (top-right). (D) Hit selection dialog showing various optional criteria. These can be used separately or combined, allowing to screen for a large variety of dynamic phenotypes. (E) Based on the criteria, hit cells are determined and visualized through a ‘hits only’ lifetime traces plot (left). Blue vertical lines indicate the chosen hit detection time window. The hit cells are outlined with white lines on the lifetime-intensity overlay image (right). (F) Additional visualizations of hit cells (left) and non-hit cells (right), in density graphs, resp…
Beyond Static Screens: A High-Throughput Pooled Imaging CRISPR Platform for Dynamic Phenotype Discovery by Kees Jalink and team: www.biorxiv.org/content/10.1...
14.07.2025 07:35 — 👍 15 🔁 3 💬 2 📌 0
Hi everyone! I’m new to Bluesky. At the beginning of the year, I moved to Zurich @UZH_en to start a postdoc with #CyrilZipfel and @pedrobeltrao.bsky.social, investigating protein phosphorylation in plants 🌱
If you’re nearby or working on something similar, let’s connect 😀

🌍Euromit 2026 is coming to Angers!🌍
✨The international congress dedicated to mitochondrial research, will take place in France, at the Centre des Congrès of Angers. from May 31 to June 4, 2026! ✨
📢 Stay informed by following this page and get ready for an exceptional scientific experience! 🚀🔬

Impairing mitochondrial fusion triggers a vicious cycle of unequal mitochondrial inheritance and failed mtDNA synthesis that rapidly depletes mtDNA:
www.biorxiv.org/content/10.1...

Origin and evolution of mitochondrial inner membrane composition
journals.biologists.com/jcs/article/...
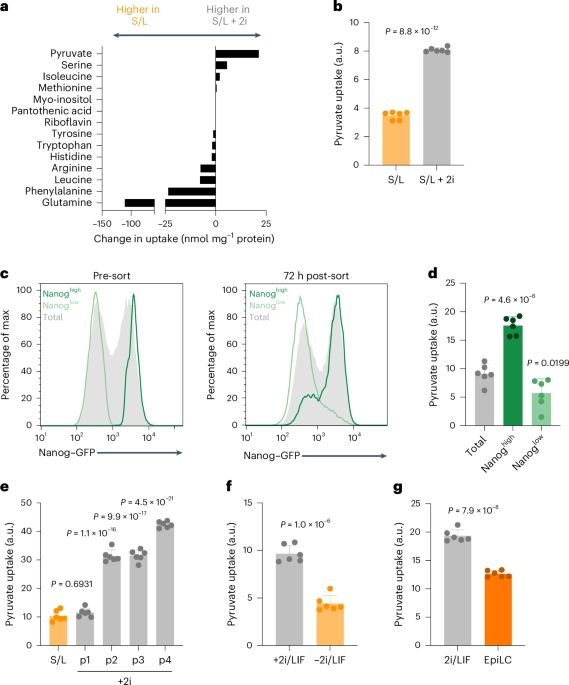
Intracellular metabolic gradients dictate dependence on exogenous pyruvate
www.nature.com/articles/s42...
Congrats to @lydiafinley.bsky.social Lab!
Very cool work from the @katajistolab.bsky.social ! Congratulations to the team!
29.04.2025 06:50 — 👍 6 🔁 1 💬 1 📌 0
Bluesky has overtaken its flailing rival X in hosting posts related to new academic research, indicating the platform is fast becoming the go-to place for scholars to share their work
#AcademicSky #EduSky
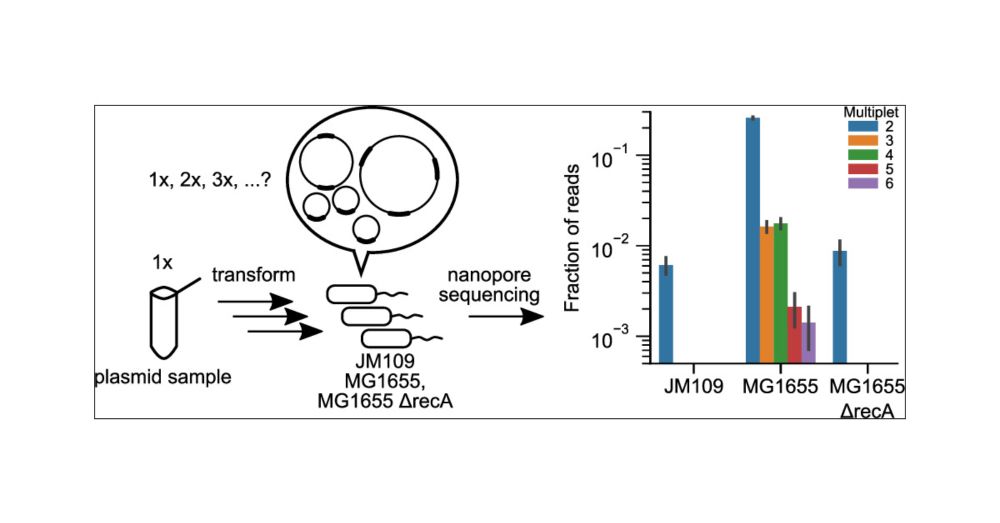
Preventing Multimer Formation in Commonly Used Synthetic Biology Plasmids | ACS Synthetic Biology pubs.acs.org/doi/full/10....
24.03.2025 09:40 — 👍 2 🔁 0 💬 0 📌 0
Check out our new fluorescent probe for imaging f-actin dynamics: 𝗦𝗶𝗥-𝗫𝗔𝗰𝘁𝗶𝗻: 𝗔 𝗳𝗹𝘂𝗼𝗿𝗲𝘀𝗰𝗲𝗻𝘁 𝗽𝗿𝗼𝗯𝗲 𝗳𝗼𝗿 𝗶𝗺𝗮𝗴𝗶𝗻𝗴 𝗮𝗰𝘁𝗶𝗻 𝗱𝘆𝗻𝗮𝗺𝗶𝗰𝘀 𝗶𝗻 𝗹𝗶𝘃𝗲 𝗰𝗲𝗹𝗹𝘀 www.biorxiv.org/content/10.1...
Thank you @veselin-nasufovic.bsky.social
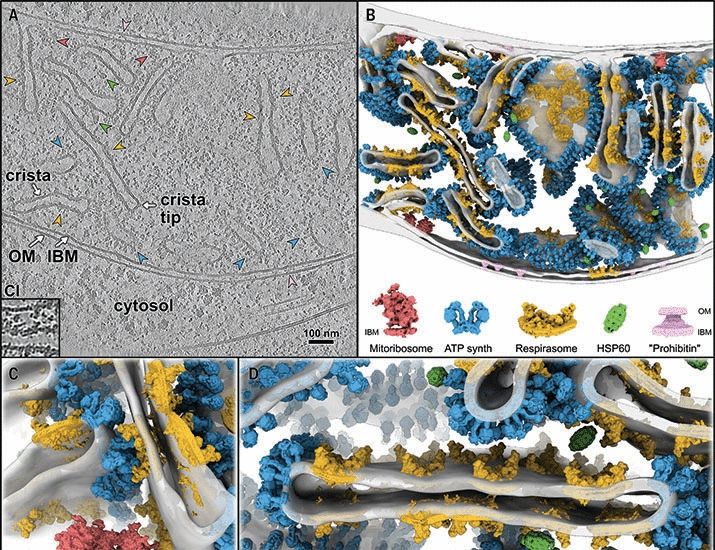
In-cell architecture of the mitochondrial respiratory chain | Science www.science.org/doi/10.1126/...
21.03.2025 09:33 — 👍 33 🔁 13 💬 0 📌 0
Join us at the ICSB in Dublin this October - I will chair the session on aging systems biology
icsb2025.com

Interesting preprint about ATP gradients within cells: www.biorxiv.org/content/10.1...
21.02.2025 11:54 — 👍 3 🔁 1 💬 0 📌 0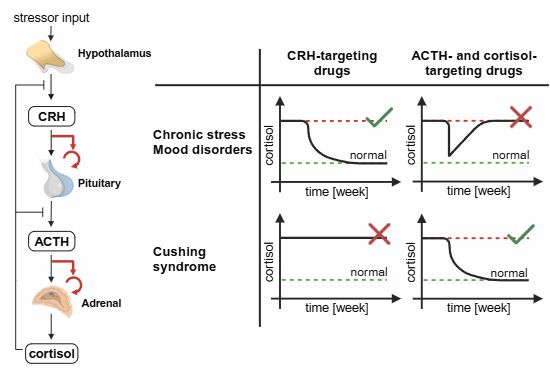
Why do cortisol-lowering drugs work for Cushing’s but fail in mood disorders & chronic stress?
We propose that the ability of the pituitary and adrenal glands to adjust their functional masses—normally beneficial—counteracts the drug’s effect. Using a math. model, we screened HPA interventions (1/2)
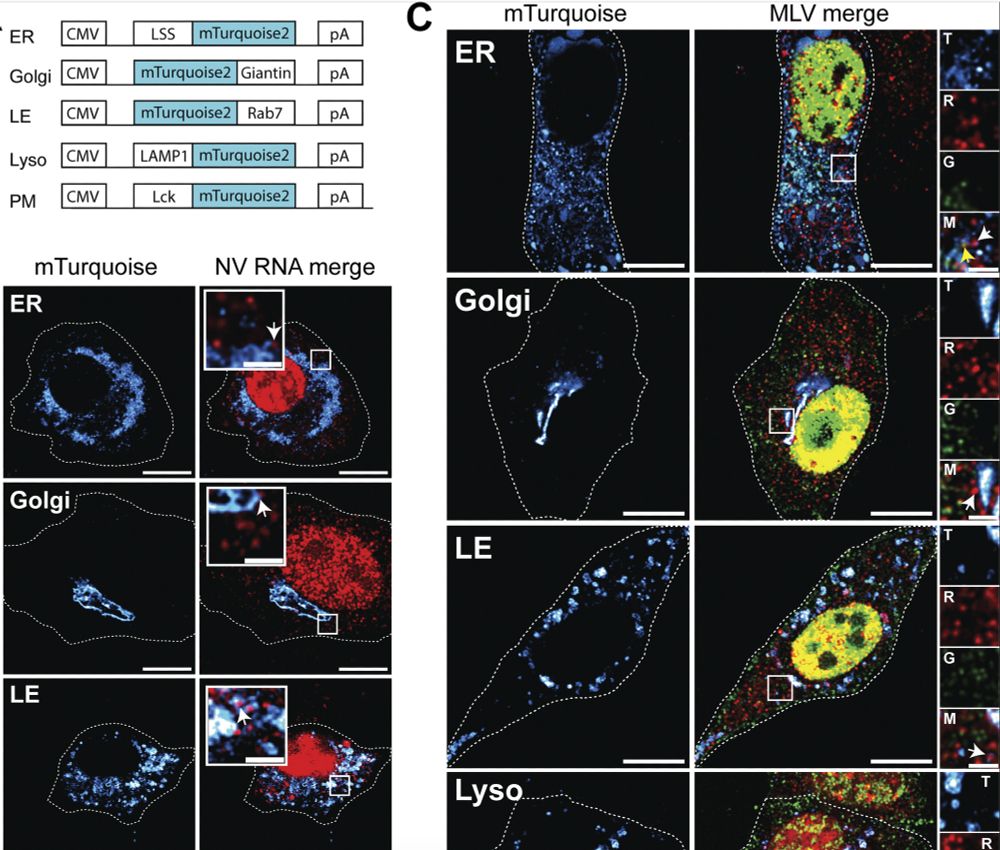
Subcellular localization of non-translating (free) RNAs and translation sites. (A) Schematic maps of the mTurquoise markers used to label different subcellular sites. (B) Representative images of the NIH3T3 GFP/RFP cells co-transfected with NV-MS2 and one of each mTurquoise plasmid labeling ER, Golgi, LE, Lyso, and PM. Images were acquired on an LSM980 confocal microscope. RNA is in red and subcellular locations in hot cyan. Single z-planes are presented, and scale bars are 10 and 2 µm for zoom insets. White arrows indicate NV RNA associated with the turquoise surfaces. (C) Representative images of cells co-expressing ΔNC and one of each mTurquoise plasmid. Translating and non-translating FL RNAs associated with the subcellular surfaces (hot cyan) are indicated by yellow and white arrows, respectively. Single z-planes are presented, and scale bars are 10 and 2 µm for zoom insets. In zoom insets, the letters T, R, G, and M denote turquoise, red, green, and merge, respectively.
Great to see the mTurquoise2 plasmids that we've generated used as markers of subcellular structures
rupress.org/jcb/article/...
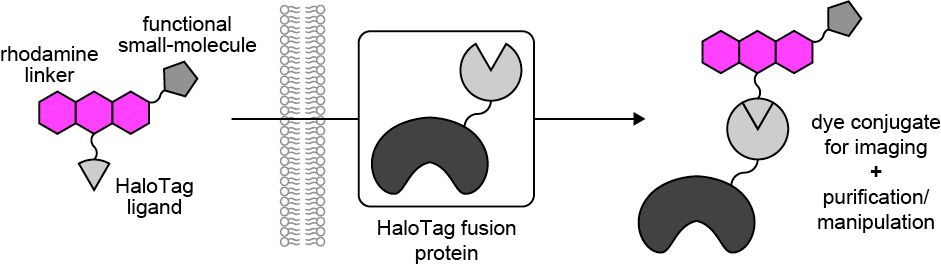
🧵Prepint alert! Optimizing Multifunctional Fluorescent Ligands for Intracellular Labeling | tinyurl.com/3n55hvsc. With Jason Vevea, Ed Chapman, and @so-lets-kilab70.bsky.social, we combined dye chemistry, HaloTag, microscopy and cell biology to make protein purification and manipulation tools.
28.01.2025 05:17 — 👍 65 🔁 25 💬 2 📌 2Mito-friends:
What conferences are we attending this year?
I ask because there are no mito/cyto symposia at my usual conferences, and it would be great to catch up!
🔬🧪

Our global study on the state of trust in scientists is now out in Nature Human Behaviour! 🥳
With a team of 241 researchers, we surveyed 71,922 people in 68 countries, providing the largest dataset on trust in scientists post-pandemic 👇🧵https://www.nature.com/articles/s41562-024-02090-5
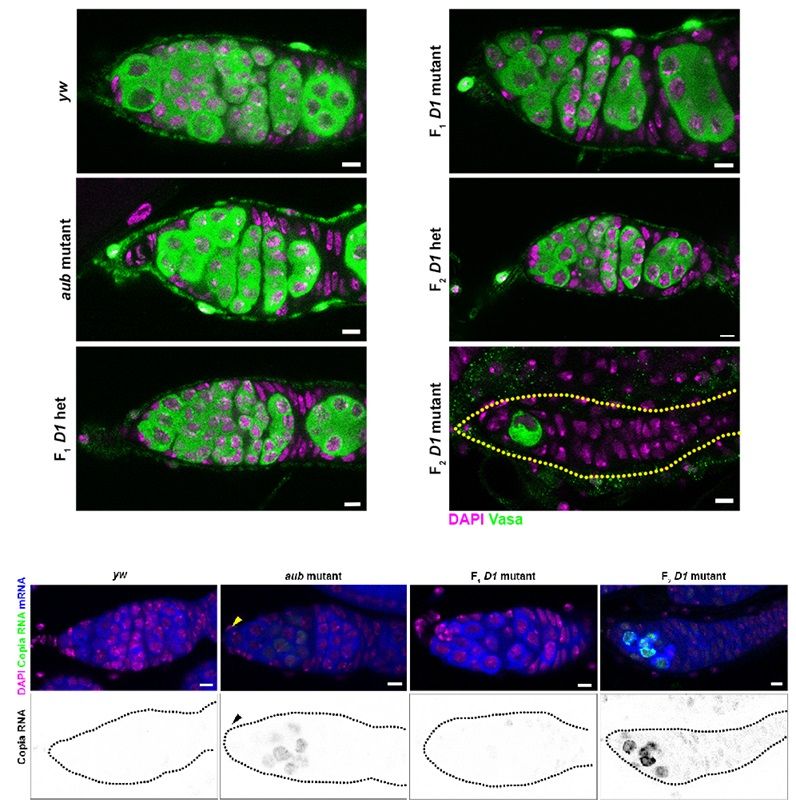
Loss of D1 during embryogenesis triggers transposon expression in adult ovaries. Top: Ovaries from the indicated genotypes (D1 and aubergine mutants and heterozygotes, F1 and F2) were stained with Vasa (green) and DAPI (magenta). Bottom: RNA in situ hybridization against Copia (green) and total polyadenylated mRNA (blue) was performed in ovaries of the indicated genotypes that were also stained for DAPI (magenta). Yellow and black arrowheads indicate undifferentiated germ cells in the aubergine mutant germarium.
Pericentromeric #heterochromatin clusters into chromocenters. @skrutlnize.bsky.social &co use quantitative mass spec to characterize the chromocenter proteome in multiple tissues, identifying a link between satellite DNA-binding proteins & #transposon repression🧪 @plosbiology.org plos.io/4akuNAi
16.01.2025 13:13 — 👍 14 🔁 10 💬 1 📌 0FLIM imaging reveals IMM lipid heterogeneity in live cells.
Find out more about this exciting tool: onlinelibrary.wiley.com/doi/10.1002/...
Congratulations on that cool story! I like the short descriptive titles in your figures.
07.01.2025 16:58 — 👍 1 🔁 0 💬 1 📌 0
Stoked to share TWO preprints on mitochondrial pearling! 🎉
We uncover how spontaneous #mitochondria pearling drives #mtDNA nucleoid distribution (doi.org/10.1101/2024...), and
@gavsturm.bsky.social
et al. dissect the biophysics behind it (doi.org/10.1101/2024...). Details below! 🔬🧵🧪
This is fascinating, congratulations!
07.01.2025 16:24 — 👍 0 🔁 0 💬 0 📌 02x Preprint drop!!! Mitochondrial pearling has arrived!
“The Biophysical Mechanism of Mitochondrial Pearling”.
“Pearling Drives Mitochondrial DNA Nucleoid Distribution” 1/10
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
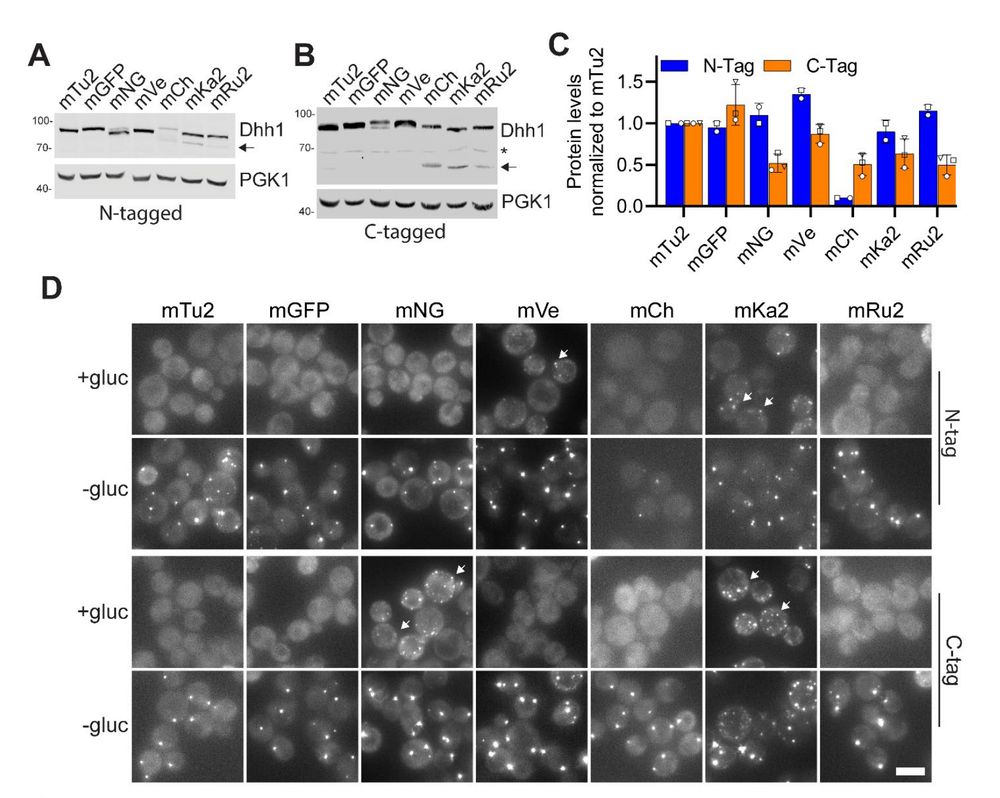
Figure 4. Position and type of fluorescent tag affect protein expression and P-body numbers in vivo.
The dark side of fluorescent protein tagging: the impact of protein tags on biomolecular condensation
www.biorxiv.org/content/10.1...
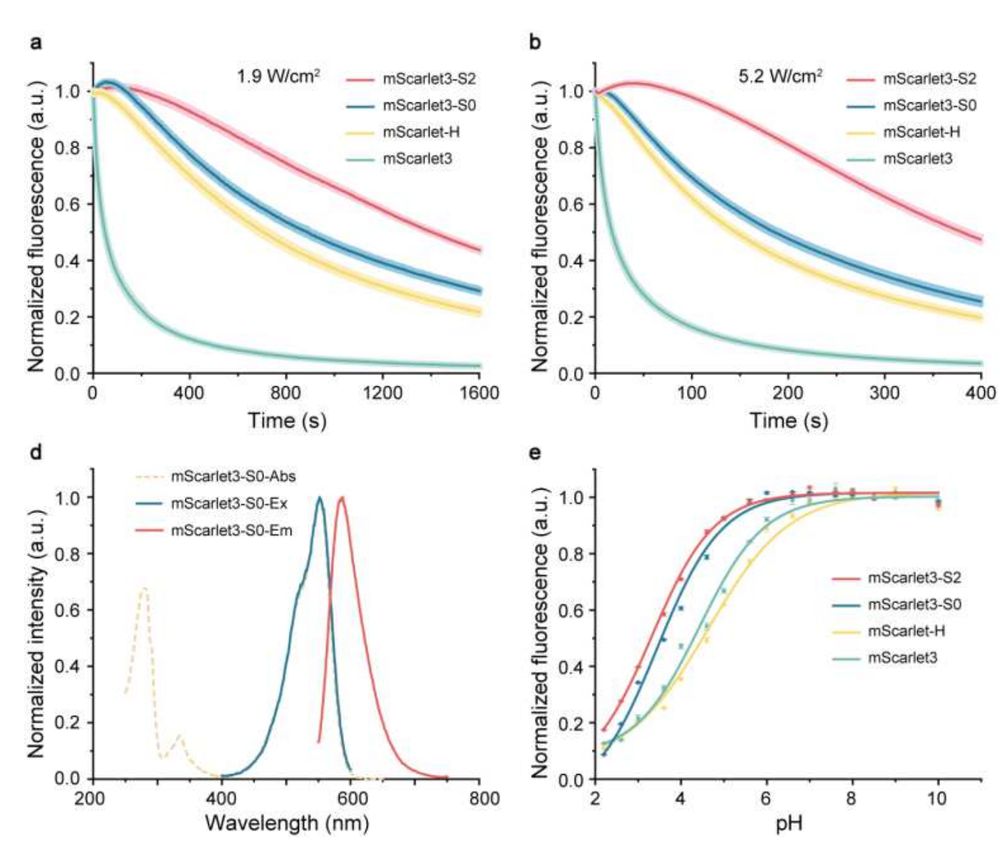
Fig. 1 Photostable and fluorescence properties of mScarlet3-S2. a, b, Photostability of red emitting mScarlet variants expressed in live U-2 OS cells under continuous WF illumination at 1.9 W/cm2
A highly photostable monomeric red fluorescent protein
www.researchsquare.com/article/rs-5...
Interesting article:
rupress.org/jcb/article/...
Vimentin-mSG FRAP
17.12.2024 05:58 — 👍 80 🔁 11 💬 2 📌 1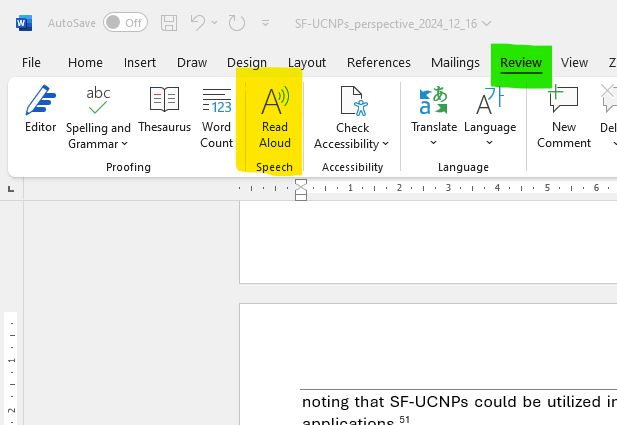
Struggling with proof-reading? A reminder that MS Word can read your document back to you via the 'read aloud' funciton found in the 'review' tab. An absolute game changer!
17.12.2024 12:09 — 👍 7 🔁 2 💬 1 📌 0