Quote from Dr. Mingda Ye, University of Oxford, highlighting the collaborative breakthrough in cryo-EM imaging of small proteins: “This idea was initiated when solving sub-50kDa protein structures by cryo-EM was almost impossible. To break this barrier, many world-class scientists in different fields have combined forces and it is a great honour to work with them to bring this game-changing tool into reality!” The background diagram outlines the scientific workflow used in the study.
Exciting work from the Franklin, @ox.ac.uk, and @diamondlightsource.bsky.social has led to a new method for imaging small proteins (<50 kDa) using cryoEM. By using bifunctional, bispecific nanobody scaffolds, the team have successfully solved the smallest protein structure to date (14 kDa).
18.08.2025 15:27 — 👍 13 🔁 8 💬 1 📌 0

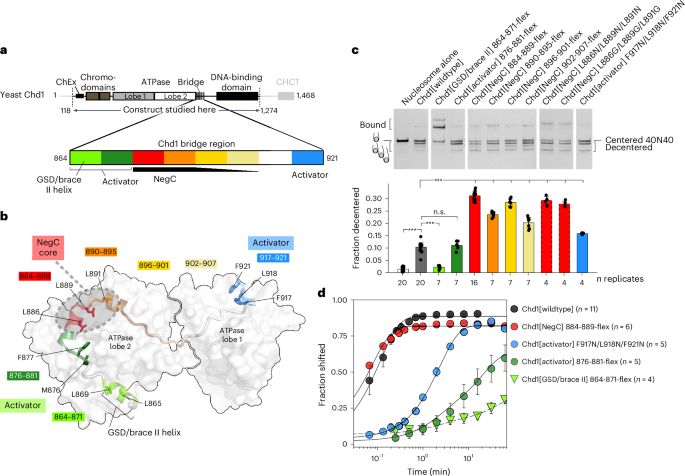
Collagen VI microfibril structure reveals mechanism for molecular assembly and clustering of inherited pathogenic mutations
Nature Communications - Collagen VI microfibril cryo-EM structure resolves a cysteine-rich coiled-coil important for heterotrimerization and microfibril assembly, reveals a hotspot of collagen VI...
Amazing new paper from Clair Baldock's lab resolves the collagen VI microfibril structure by cryo-EM to reveal a cysteine-rich coiled-coil crucial for heterotrimerization & microfibril assembly. The structure also reveals a hotspot of collagen VI muscular dystrophy mutations that disrupt assembly.
15.08.2025 06:31 — 👍 2 🔁 1 💬 0 📌 0
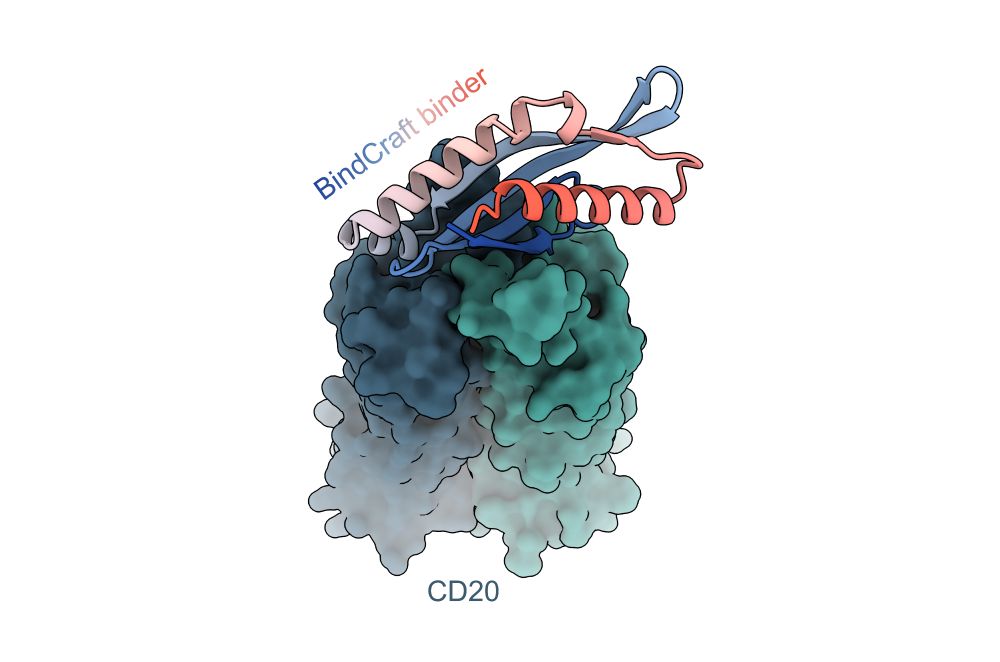
We have written up a tutorial on how to run BindCraft, how to prepare your input PDB, how to select hotspots, and various other tips and tricks to get the most out of binder design!
github.com/martinpacesa...
30.06.2025 19:45 — 👍 138 🔁 55 💬 4 📌 0
🚀 Excited to release BoltzDesign1!
✨ Now with LogMD-based trajectory visualization.
🔗 Demo: rcsb.ai/ff9c2b1ee8
Feedback & collabs welcome! 🙌
🔗: GitHub: github.com/yehlincho/Bo...
🔗: Colab: colab.research.google.com/github/yehli...
@sokrypton.org @martinpacesa.bsky.social
03.06.2025 01:30 — 👍 54 🔁 17 💬 1 📌 0
I also had fun in making gummy GLT25D1 making #notthecover using #blender
22.04.2025 09:33 — 👍 3 🔁 1 💬 0 📌 0
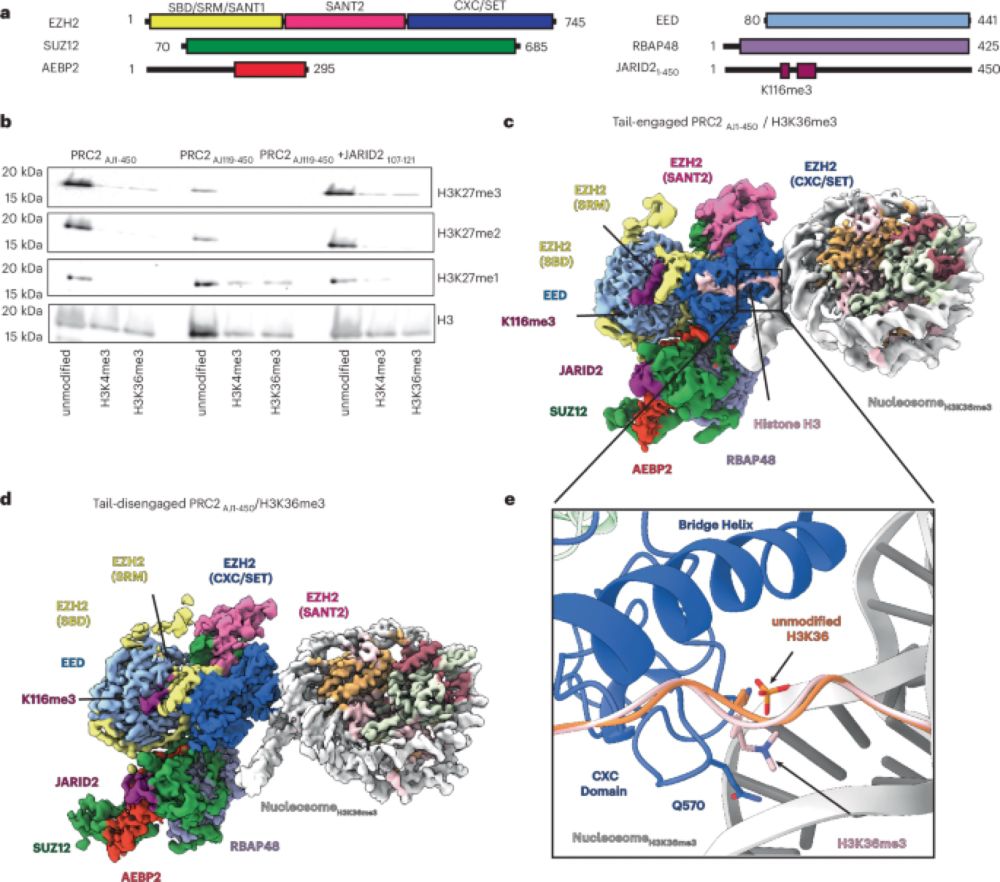
This was a massive work from @fornerislab.bsky.social fellows to ehihc I contributed when I was Postdoc, and great #teamwork with Giorgio Colombo lab, @claudio-iacobucci.bsky.social and Alberta Pinnola lab.
22.04.2025 09:33 — 👍 1 🔁 1 💬 1 📌 0
we found that the GT1 domain, while not catalytic, is surprisingly capable of binding Ca²⁺ and UDP-α-galactose. This was intriguing to us, but all the mutants in the GT1 site did not produce folded protein, consistent with a structural role for the GT1 (also confirmed by MD).
22.04.2025 09:33 — 👍 0 🔁 0 💬 1 📌 0
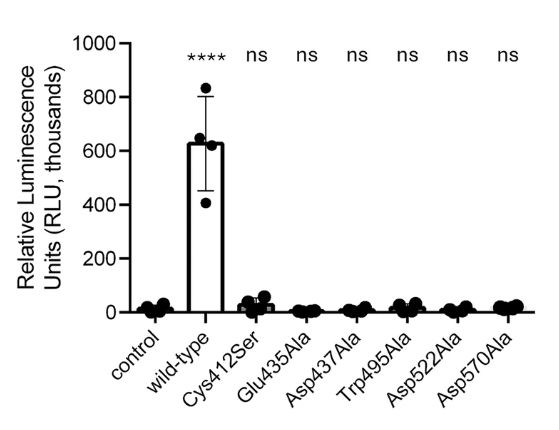
And here the question: are they both active? which one is the responsible for the GalT activity? We combined #mutagenesis with #biophysics and #moleculardynamics to show that only the GT2 domain exhibits catalytic activity, facilitated by an unusual Glu-Asp-Asp motif critical for Mn²⁺ binding.
22.04.2025 09:33 — 👍 1 🔁 1 💬 2 📌 0
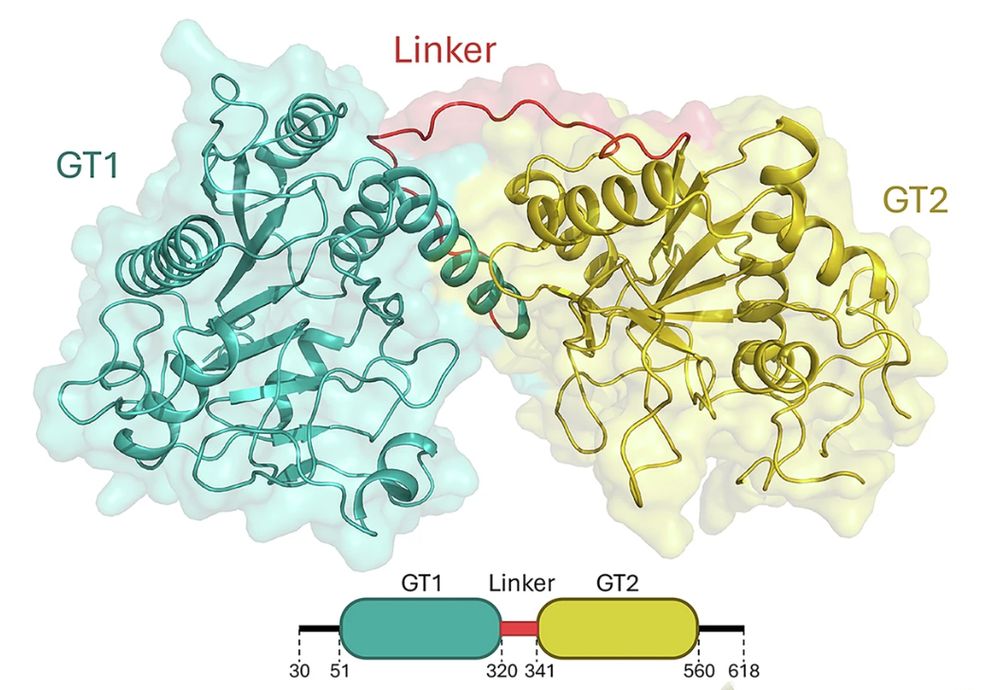
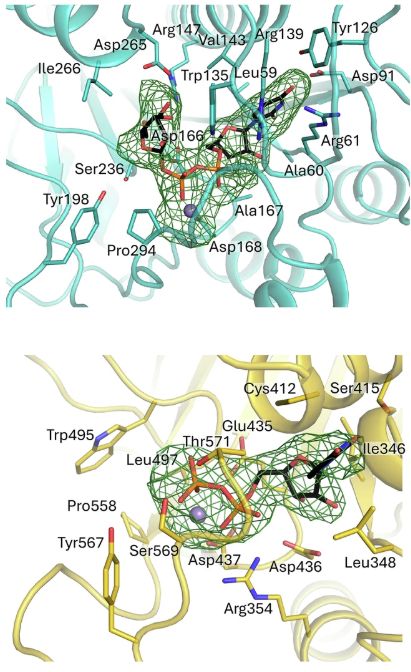
Each monomer contains two domains (GT1 and GT2) connected by a long but ordered linker. Surprisingly, both domains are capable of binding metal ions and the donor substrate UDP-α-galactose.
22.04.2025 09:33 — 👍 0 🔁 0 💬 1 📌 0
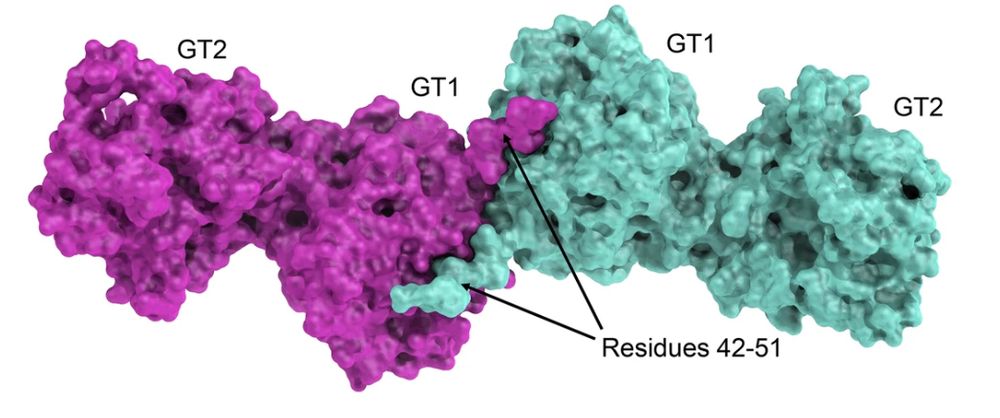
We solved the #Xray structure of GLT25D1/COLGALT1 via experimental phasing (yes, it was before #Alphafold) showing that it forms an elongated head-to-head homodimer
22.04.2025 09:33 — 👍 1 🔁 0 💬 1 📌 0
Collagen's function are ensured by essential post-translational modifications including galactosylation.
GLT25D1 is the galactosyltransferase enzyme responsible for initiating the glycosylation of hydroxylysine in collagen. but its structure was missing until now
22.04.2025 09:33 — 👍 0 🔁 0 💬 1 📌 0
Super happy to share the final shape of our work describing the molecular #structure and #enzymatic mechanism of human #collagen #galactosyltransferase #GLT25D1 published in @natcomms.nature.com. Want to know how collagen become sweet? read the 🧵 below!
22.04.2025 09:33 — 👍 4 🔁 1 💬 1 📌 0
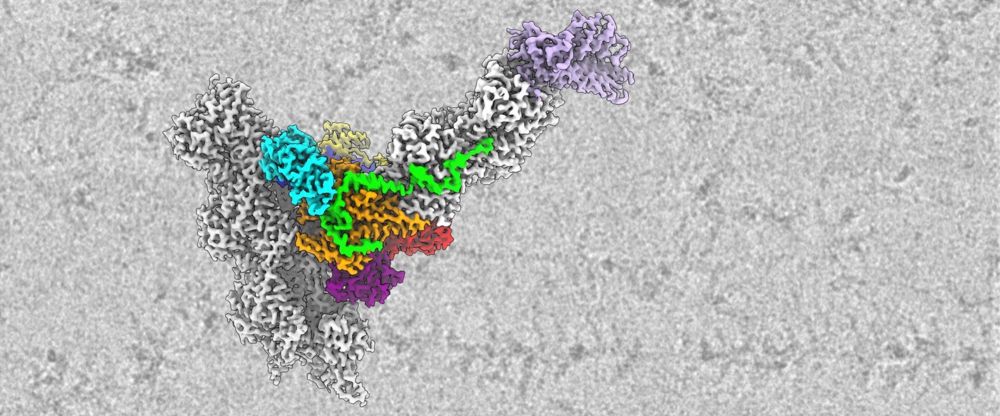
Image processing for cryo-electron microscopy
Cryo electron microscopy (cryo EM) is a major structural biology method for studying macromolecular complexes and cellular structures in their native states. Stable high resolution cryo microscopes, …
We are thrilled to announce the 2025 EMBO practical course in cryo-em image processing, Birkbeck College, London, 9-16 Sept 2025. More info & apply: meetings.embo.org/event/25-cry... Organisers Giulia Zanetti & Helen Saibil. Beautiful image from co-organizer @carolynmoores1.bsky.social lab.
18.03.2025 19:06 — 👍 31 🔁 18 💬 1 📌 4
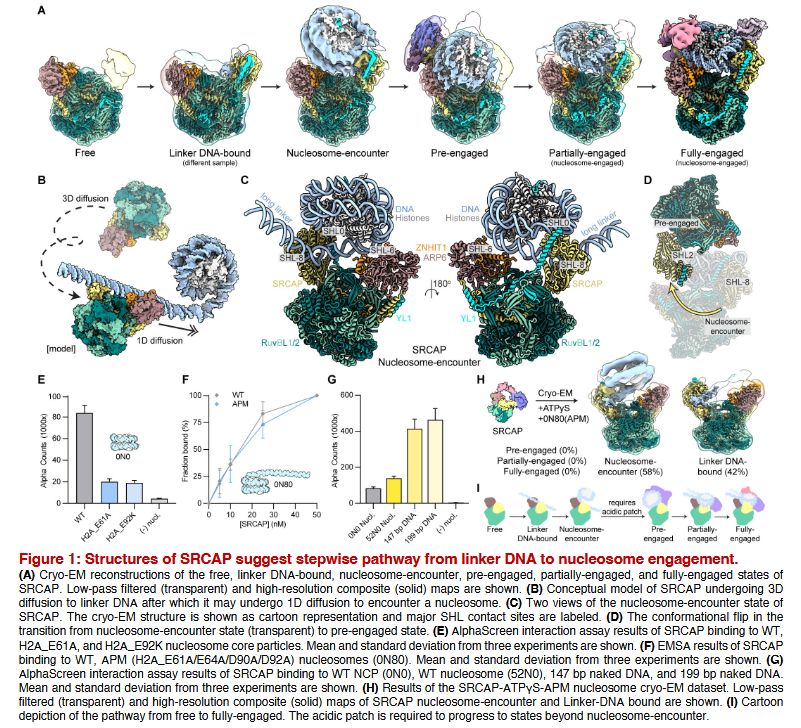
This is a masterpiece ! #cryoEM
Structures of H2A.Z-associated human chromatin remodelers SRCAP and TIP60 reveal divergent mechanisms of chromatin engagement
www.biorxiv.org/content/10.1...
04.03.2025 08:47 — 👍 20 🔁 3 💬 0 📌 0
Did you make our cryo-EM webinar last week? Dive into pre-processing, how to interpret 2D classes, 3D reconstructions from the initial map to final structure with CSO Giovanna Scapin.
Catch it here: youtu.be/2dBnPVkaoFs
Sign up for Overcoming Limitations, on 3/25: nimgs.zoom.us/webinar/regi...
04.03.2025 18:34 — 👍 4 🔁 1 💬 0 📌 0
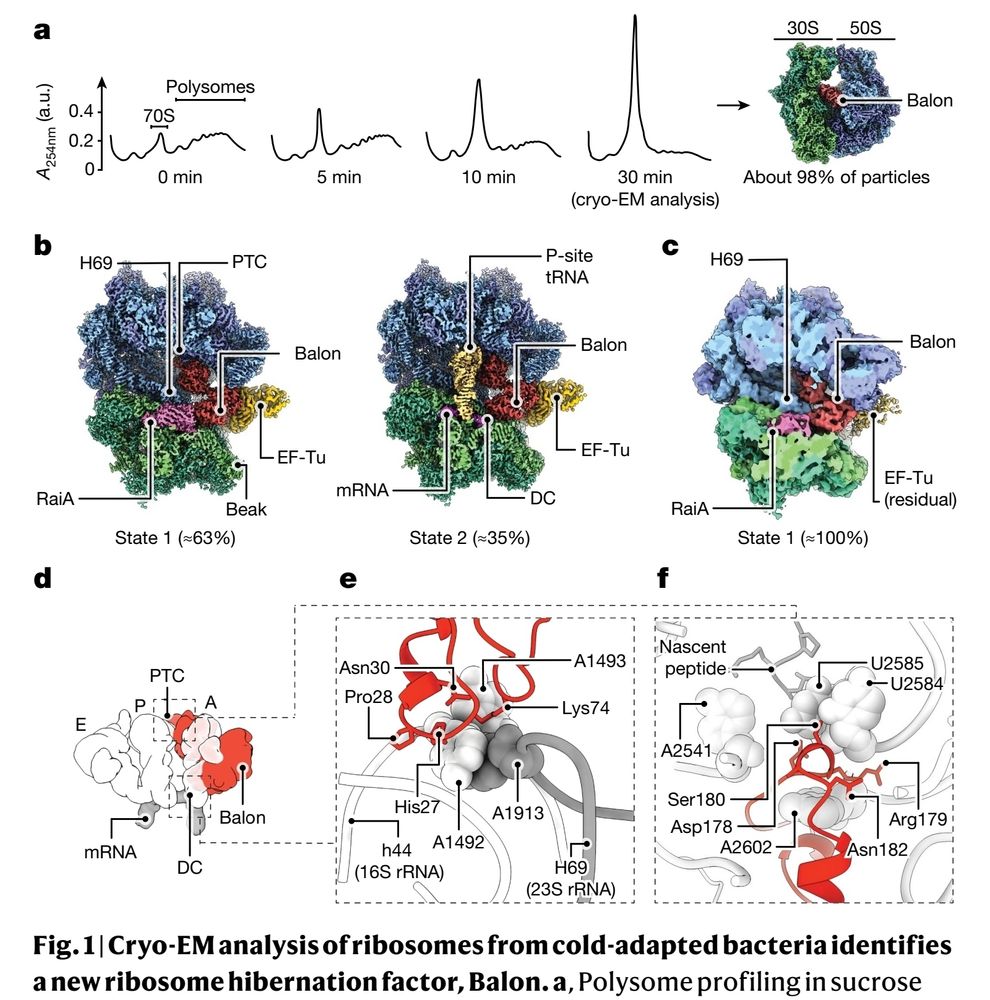
Been thinking about creating a collection of good protein structure figures, as inspiration for my own work.
#1 www.nature.com/articles/s41...
03.03.2024 08:37 — 👍 167 🔁 54 💬 7 📌 10
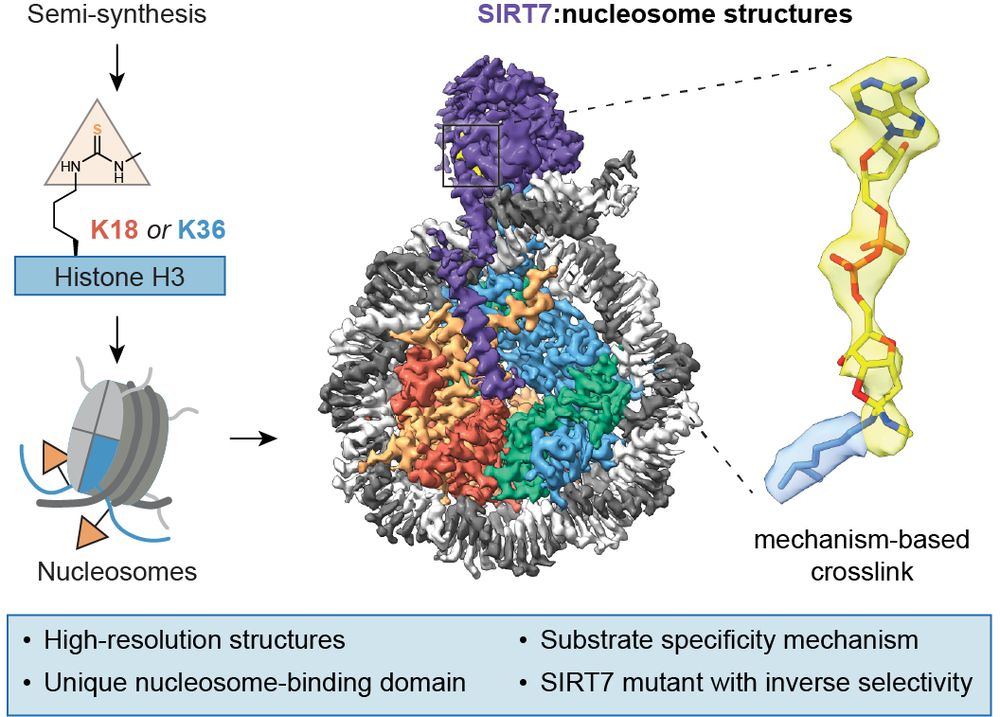
#SIRT7 is a histone deacetylase with highly specific activity on #chromatin substrates.
We just published mechanism-based #cryoEM structures of #SIRT7 on nucleosomes to understand its activity 👇
www.nature.com/articles/s41...
(1/8) #ChemBio #ChemSky
04.02.2025 13:13 — 👍 43 🔁 17 💬 1 📌 1
Cool event, great people! Thanks for putting us together @fornerislab.bsky.social and @thermofishersci.bsky.social!
29.01.2025 21:00 — 👍 1 🔁 0 💬 0 📌 0
Exciting new preprint alert! 🚨 The Collepardo lab, teaming up with Huabin Zhou and the Rosen lab we took a deep, high-resolution look into chromatin condensates! [1/6] #Chromatin #MD @rcollepardo.bsky.social @juliamaristany.bsky.social
www.biorxiv.org/content/10.1...
22.01.2025 17:31 — 👍 37 🔁 12 💬 1 📌 3
Truly honoured to be the first user of such an amazing platform! For sure a reference point for all the Italian community. Exciting time ahead! ❄️🚀
14.01.2025 10:17 — 👍 6 🔁 0 💬 0 📌 0
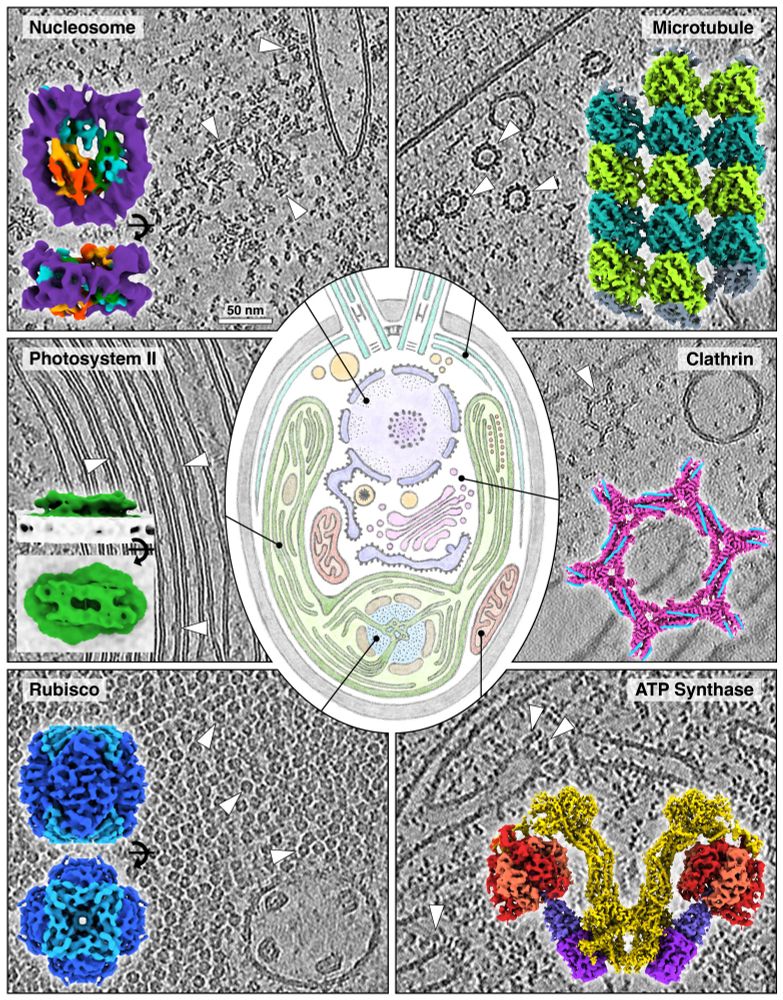
Towards community-driven visual proteomics! Excited to finally share this large-scale curated & annotated dataset of 1829 high-quality #cryoET tomograms of the little green alga that just keeps giving— Chlamydomonas! 🧪🧶🧬🌾🌊🌍
Preprint📜: www.biorxiv.org/content/10.1...
A short thread🧵👇
06.01.2025 11:18 — 👍 337 🔁 128 💬 7 📌 14
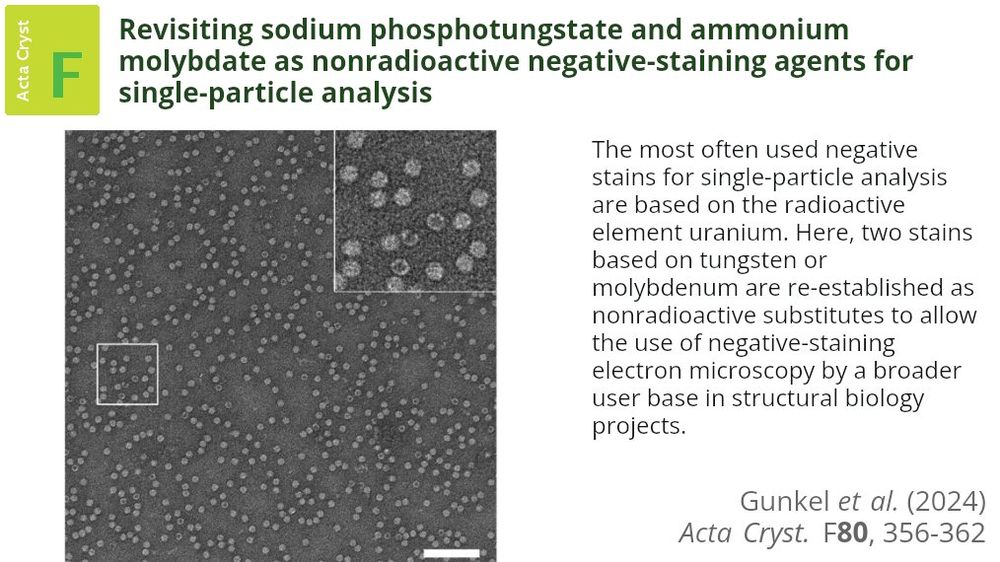
Safer, nonradioactive staining alternatives for electron microscopy are introduced, simplifying sample preparation, reducing costs and making structural biology more accessible to researchers globally #NegativeStaining #ElectronMicroscopy doi.org/10.1107/S205...
17.12.2024 17:13 — 👍 16 🔁 6 💬 0 📌 0
Structural biologist moved to the dark side of comp bio & science visualization 🧪 @BioRender | SciComm & SciArt @proteinimaging | alumn @scrippsresearch
Postdoctoral researcher at the Institute of Cancer Research, London
Cryo-EM 🔬DNA repair 🧬
Associate Professor interested in molecular glycobiology, glycomics of lower eukaryotes, glycan arrays, chemical glycobiology, classical music. Viva la libertà!
Ph.D. in Vannini lab @HumanTechnopole
PhD student in Structural Biology at Human Technopole, Milan, Italy. Passionate about RNA related world.
Predoctoral Research Assistant at the Llorca lab @cniostopcancer.bsky.social working on mTORC1 signaling and cryo-EM ❄️🔬. Intrigued by the structural mechanisms of protein assemblies involved in endomembrane signaling and trafficking 🧪.
let's talk about microscopy and/or neuroscience, anything at any time!
I like large rocks and small proteins.
BIF fellow doing cryoEM+Protein Design in the Llorca lab ❄️🔬
Scientist - Structural Proteomics, Mass spectrometry at Human Technopole. Views are my own. #XL-MS #TeamMassSpec #StructuralProteomics #StructuralBiology #Proteomics
Your guide to AI-driven protein design, DNA nanotech & more! Subscribe at https://plentyofroom.beehiiv.com/
Glycobioinfonaut, Associate Professor at the Copenhagen Center for Glycomics, University of Copenhagen. @hirenj.11 on Signal
Assistant Professor at the University of L'Aquila, Italy.
Led by Paola Scaffidi, we are a group at IEO and interested in Epigenetics, Chromatin and Cancer https://www.research.ieo.it/research-and-technology/principal-investigators/paola-scaffidi/
Structural biologist working on 🖥️ protein design, machine learning🤖, crystallography💎, and cryoEM🔬. Avid weirdness connoisseur 🎩
Working on self organization, cilia, and IFT. Cell biologist, CLEM and cryo-EM expert at Human Technopole. Was a serious fencer in a previous life. Love pottery!!! 🏺
@DamonRunyon Postdoc Fellow in Tim Springer lab @harvardmed and @BostonChildrens | Organoids, Integrins, Antibodies, CryoEM | @Cornell & @IISERPune alumnus | He/Him