Thank you @stefanschoenfelder.bsky.social - far too modest!
07.06.2025 21:25 — 👍 1 🔁 0 💬 0 📌 0

Flynn Lab
FLYNNLAB
Chromatin biology
Many thanks especially to Colm Doyle (now at @embl.org), who began this work with his beta globin locus study and @seanflynn.bsky.social (now of sites.google.com/view/flynnlab/ at @exeter.ac.uk - great things to come) for an incredible journey, as well as all authors at @cruk-ci.bsky.social! 9/9
04.06.2025 21:00 — 👍 0 🔁 0 💬 0 📌 0
....then edited them out to show how the regulation of genes in 3D contact was affected. 8/9
04.06.2025 21:00 — 👍 0 🔁 0 💬 1 📌 0
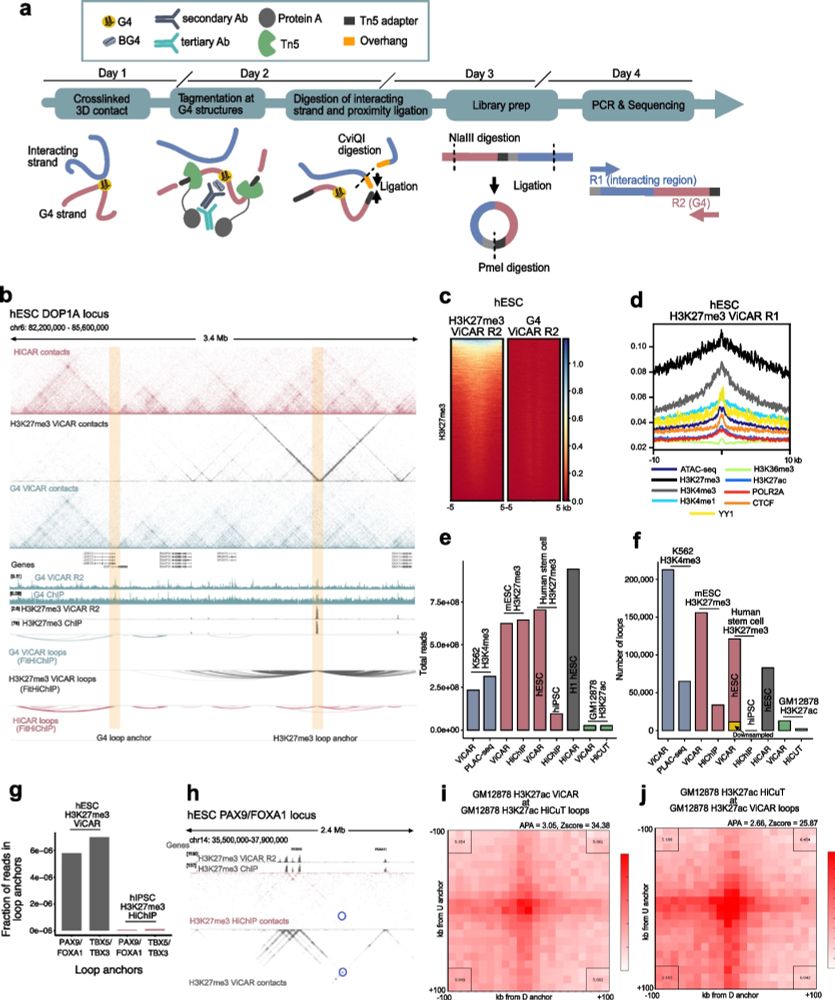
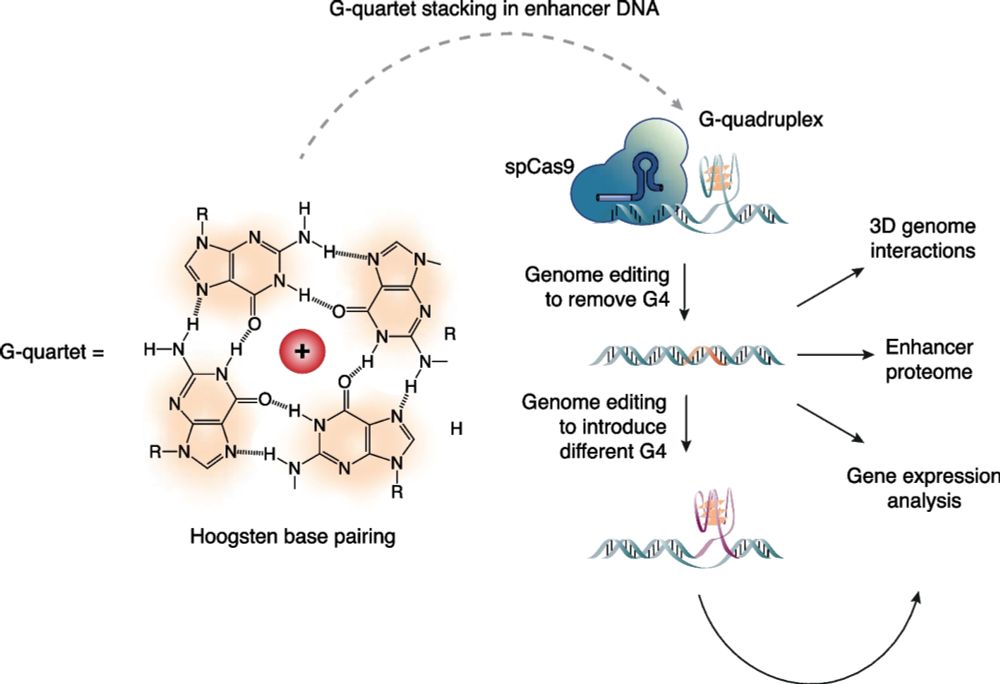
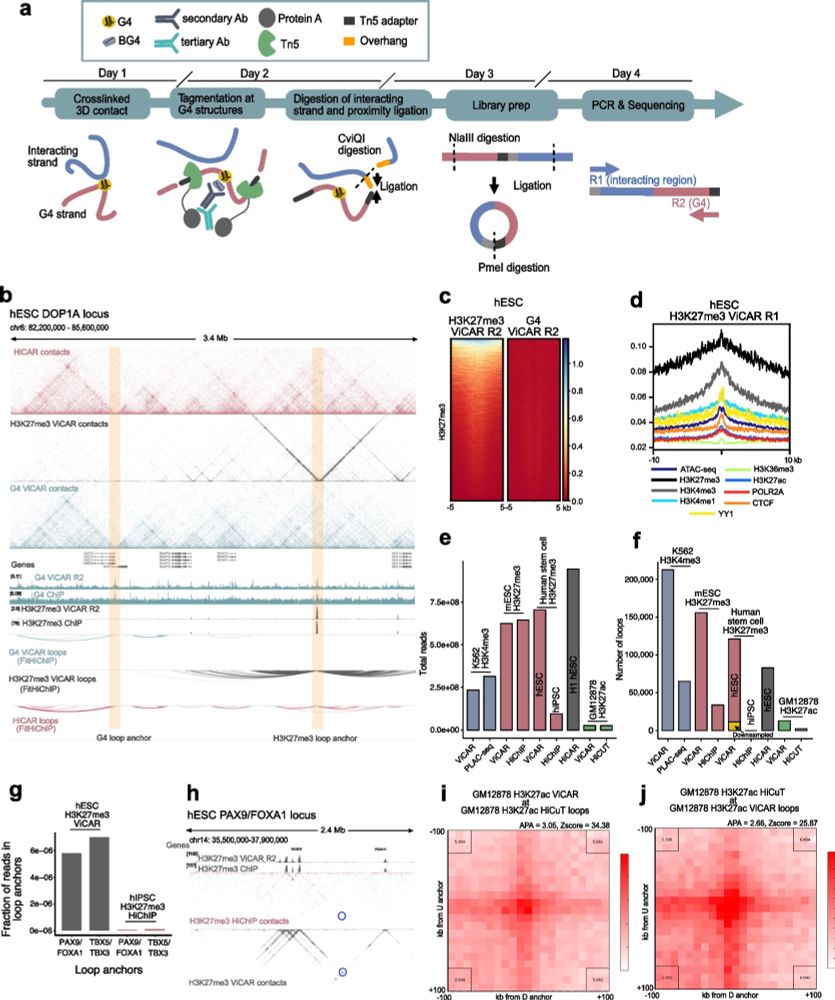
How do G4s function at enhancers? In our #proteomics pulldowns, RNA polymerase II subunits were enriched, together with known G4 binding proteins - but not some common architectural factors (e.g. YY1, CTCF) - we confirmed that some RNAPII subunits specifically bind G4s (Fig. 4A, F). 6/9
04.06.2025 21:00 — 👍 0 🔁 0 💬 1 📌 0
For the beta HS2 enhancer, this effect is masked by hyper activation of the neighbouring enhancer, HS3 - removing HS3 unveils a similar effect (Figure 3A-D for alpha, Figure 3E-H for beta). 4/9
04.06.2025 21:00 — 👍 0 🔁 0 💬 1 📌 0
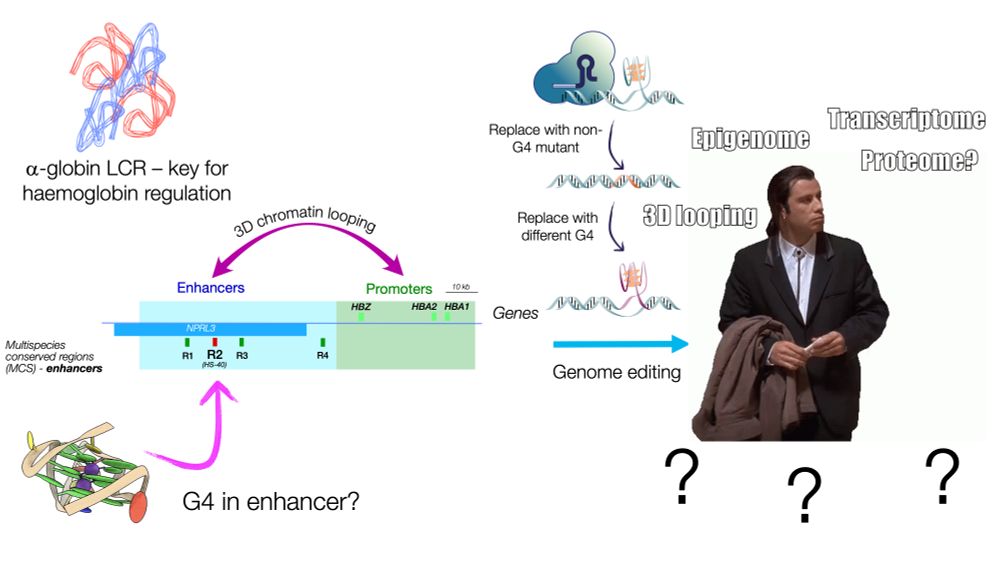
We found a G4 in HS-40, the dominant enhancer of the alpha-globin locus regulatory region, replaced it with a sequence that can no longer form a G4; cognate gene expression, chromatin accessibility, and enhancer-promoter contacts were reduced. We then introduced a different G4-forming sequence - enhancer activity was restored.
What happened? In the alpha HS-40 enhancer, such a mutation (seven G-to-T mutations) was sufficient to reduce globin gene expression to levels observed when the whole enhancer region (of around 350 bases) is deleted. Enhancer-promoter contacts and local #chromatin accessibility is also reduced. 3/9
04.06.2025 21:00 — 👍 0 🔁 0 💬 1 📌 0
Using the classic #globin #superenhancer systems, we discovered two G4 structures in dominant enhancers in both the alpha globin and beta globin locus control systems - and replaced them with non-G4-forming sequences using CRISPR. 2/9
04.06.2025 21:00 — 👍 0 🔁 0 💬 1 📌 0

We heard about "Alternative DNA structures in enhancer-promoter communication" from @seanflynn.bsky.social from @uniofexeter.bsky.social #Epigenetics2025
@uniofexeternews.bsky.social
23.04.2025 14:00 — 👍 7 🔁 2 💬 0 📌 0

🚨 We’re Hiring!🚨
Passionate about cancer research? Join Madzia Crossley's team to investigate how R-loop dysregulation contributes to cancer.
We have two exciting opportunities:
Research Associate: jobs.cam.ac.uk/job/50675/
Senior Scientific Associate: jobs.cam.ac.uk/job/50689/
19.03.2025 10:18 — 👍 7 🔁 6 💬 0 📌 1
A great opportunity to work on #epigenetics, #genetics and gene regulation in #neuroscience with an inspirational young PI - please share and apply!
03.03.2025 12:17 — 👍 1 🔁 0 💬 0 📌 0

Dr Isabel Esain Garcia receives Cancer Research UK Cambridge Institute 2024 PhD Thesis Prize
Congratulations to @isabelesain.bsky.social, who is awarded the 2024 CRUK Cambridge Institute PhD Thesis Prize.
In her doctoral research, Bel discovered a new molecular mechanism in cancer gene regulation opening up a new therapeutic window for cancer treatments.
www.cruk.cam.ac.uk/news/bel-esa...
03.12.2024 13:20 — 👍 16 🔁 3 💬 1 📌 1
A great #PhD opportunity for anyone interested in #epigenetics in #neurons and the broader context of #neuroscience - having worked with @seanflynn.bsky.social and Jack, they are brilliant choices for mentors!
22.11.2024 18:44 — 👍 7 🔁 1 💬 0 📌 0
Thank you @seanflynn.bsky.social - looking forward to sharing science (mostly #epigenetics, #G4, gene regulation and #chromatin) with everyone, especially great work from the Flynn Lab!
15.11.2024 11:55 — 👍 1 🔁 0 💬 1 📌 0
Assistant Member in csBio at Memorial Sloan Kettering. Perturb-seq, single-cell functional genomics, and techniques for perturbing the genome.
Group leader @Bar Ilan University. Interested in sex determination, stem cells and 3D genome organisation. Head of the BIU transgenic unit. ERC StG 2022. Mom of 4.
Genomics & Bioinformatics Scientist at the University of Edinburgh | Head of Edinburgh Genomics | #Omics #NGS #LongReads 🧬 🖥️
Opinions my own. He/Him
Lab: https://genomics.ed.ac.uk/
LinkedIn: https://www.linkedin.com/in/javiersantoyolopez/
Studying cellular #stress responses, particularly #senescence and its impact on #immune response, #ageing and #cancer, #tumorigenesis at Cancer Research UK CI @cruk-ci.bsky.social, University of Cambridge @cam.ac.uk
Website: naritalab.com
Gene, chromatin and genome regulation in early embryos and in stem cells, with a focus on the impact of mitosis in cell identity.
A bit of 3D gene regulation, single-cell omics and transgenic models.
Born and raised in the bay of Algeciras. Enjoying Science and Flamenco at CABD, Seville.
Lab website: https://lupianezlab.github.io/Website/
Email: dario.lupianez@csic.es
Baylor College of Medicine
Assist. Prof. @unibas.ch 🇨🇭 developing single-molecule techniques to explore the nano-dynamics of proteins&Co.
https://schmid.chemie.unibas.ch/en/
We systematically elucidate the mechanisms underlying cell division and nuclear organisation and develop advanced imaging technologies
@EMBL.org
I study Chromatin structure, organization and function.
Postdoc in the Whitehouse lab, MSKCC, NYC.
Former PhD in the Pasero lab, IGH, France.
Research Professor Johns Hopkins University @jhuartssciences.bsky.social @mdc-berlin.bsky.social @mdc-bimsb.bsky.social @neurocure.bsky.social @spp2202.bsky.social
A bio-physicist turned phys-biologist,
building models and software in genome biology.
3D genome structure in mitosis | DNA repair | meiosis.
A group leader at @IMBA_Vienna.
Dad x2.
Tenured Group Leader @JohnInnesCentre, Honorary Professor @uniofeastanglia, Honorary Group Leader @BabrahamInst. Working on RNA structure functionalities
Official account for the University of Cambridge. Follow us for research, news, events and student stories from the Cambridge community 🎓📚
Using genomics & bioinformatics to decipher immunity
teichlab.org
Independent Group Leader at the MPI-IE passionate about mechanisms governing embryonic development with a focus on transposable elements🧬 Postdoc @MundlosLab 🇩🇪| PhD from @Bourchis lab 🇫🇷| She/her
https://www.julianeglaserlab.com/
Associate Professor / enhancers - 3D genome - morphogenesis
Website: https://www.unige.ch/medecine/gede/en/research-groups/999andrey
Postdoc 👩🏽🔬👩🏽💻 | Genome Replication&Stability🧬 | Institute of Cancer Research, London 📍
https://costerlab.com/
3D genome, chromatin regulation, multi-omic TechDev @ New York Genome Center 🗽, via WEHI 🇦🇺
Genome Scientist. Studies how DNA makes humans, mice, plants, microbes. At Lawrence Berkeley National Lab and Joint Genome Institute. Views are my own.