We're looking for an instructor for the algorithms course in our Bioinformatics MS program. The course assets have already been made (by me) and used in several previous offerings, but we need an instructor! If you're in the DMV area, check it out: www.linkedin.com/posts/robert...
17.10.2025 14:55 — 👍 5 🔁 3 💬 0 📌 0
#ASHG25
14.10.2025 10:41 — 👍 0 🔁 0 💬 0 📌 0
#ASHG25
14.10.2025 10:41 — 👍 1 🔁 0 💬 0 📌 0
Thanks for this. I've scheduled a slot with you. Looking forward to it.
11.10.2025 02:01 — 👍 0 🔁 0 💬 0 📌 0
As I wrap up my PhD, I’m exploring postdoctoral and industry research opportunities at the intersection of regulatory genomics and machine learning.
If you’ll be at #ASHG2025, I’d love to connect and chat!
#Genetics #Bioinformatics
08.10.2025 00:38 — 👍 1 🔁 0 💬 0 📌 1

Excited to present my poster, “A 3D Genome Framework for Causal Variant Fine-Mapping and Tissue-Specific Gene Regulation,” at @geneticssociety.bsky.social #ASHG2025 in Boston!
📌 Epigenetics Poster (Poster #8041F)
📅 Fri, Oct 17 | 2:30–4:30 PM EDT
#Genomics #Genetics #Epigenetics #3DGenome
08.10.2025 00:38 — 👍 2 🔁 0 💬 1 📌 1

Asking BlueSky for help: For a review, I am trying to accurately credit the first paper that measured pairwise 3D distances between 2 pieces of DNA on the same chromosome (or cosmid). Is Trask 1989 the first?
I know of earlier single-locus papers (1982).
www.sciencedirect.com/science/arti...
21.09.2025 18:56 — 👍 76 🔁 34 💬 5 📌 2

bioRxiv - An unbiased survey of distal element-gene regulatory interactions with direct-capture targeted Perturb-seq
New preprint from our lab!
What can we learn about the properties of gene regulatory elements by CRISPR’ing a random set of accessible sites in human cells?
Find out here: www.biorxiv.org/content/10.1...
👇
1/
19.09.2025 03:03 — 👍 58 🔁 17 💬 1 📌 1

Activity of most genes is controlled by multiple enhancers, but is there activation coordinated? We leveraged Nanopore to identify a specific set of elements that are simultaneously accessible on the same DNA molecules and are coordinated in their activation. www.biorxiv.org/content/10.1...
18.08.2025 12:23 — 👍 98 🔁 39 💬 2 📌 2
I'd love to be added! I work on computational methods for 3D regulatory genomics.
29.08.2025 00:09 — 👍 0 🔁 0 💬 1 📌 0

A 3D genome compendium of breast cancer progression
Genomics; Chromosome organization; Molecular Genetics; Cancer
We all know that cancers -even from the same subtype- are highly heterogeneous right? In a new study we asked whether this was also the case for the three-dimensional organization of the genome in breast cancer. The answer: it’s complicated. www.cell.com/iscience/ful...
18.08.2025 14:41 — 👍 14 🔁 5 💬 1 📌 0
Gene-Gene Interactions Between A LMNA Variant and Common Polymorphisms Drive Early-Onset Atrial Fibrillation https://www.medrxiv.org/content/10.1101/2025.05.05.25326834v1
06.05.2025 01:55 — 👍 2 🔁 1 💬 0 📌 0
Our study shows how combining chromatin profiling, CRISPR-based functional assays, and large-scale biobank data can advance our understanding of genetic risk in atrial fibrillation.
#AFib #LMNA #iPSC #Genomics #Preprint #RegulatoryGenomics #3DGenomics #AtrialFibrillation #Cardiogenomics
09.05.2025 03:04 — 👍 1 🔁 0 💬 0 📌 0
We applied a polygenic risk score (PRS) for AF to carriers of rare LMNA variants in UK Biobank and All of Us. Those with a high PRS had 2x higher risk of early-onset AF. This shows the value of combining rare and common variants in risk prediction. #Genomics #PRS
09.05.2025 03:04 — 👍 1 🔁 0 💬 1 📌 0
Congrats! Awesome news!
23.04.2025 13:52 — 👍 0 🔁 0 💬 1 📌 0
Effects of Lamina-Chromatin Attachment on Super Long-Range Chromatin Interactions https://www.biorxiv.org/content/10.1101/2025.02.13.638183v1
18.02.2025 04:33 — 👍 4 🔁 1 💬 0 📌 0


📢Attending @biophysicalsoc.bsky.social in LA?📢
Excited about uncovering functional links between non-coding regulatory variants and target genes? 🧬
Join me for:
🎤 Flash Talk (#1682) – Feb 18 | 12:30 PM
📊 Poster (#2754-Pos / B218) – Feb 19| 10:30 AM
See you there!
#BPS25 #Genetics #Epigenetic
16.02.2025 16:58 — 👍 1 🔁 0 💬 0 📌 0
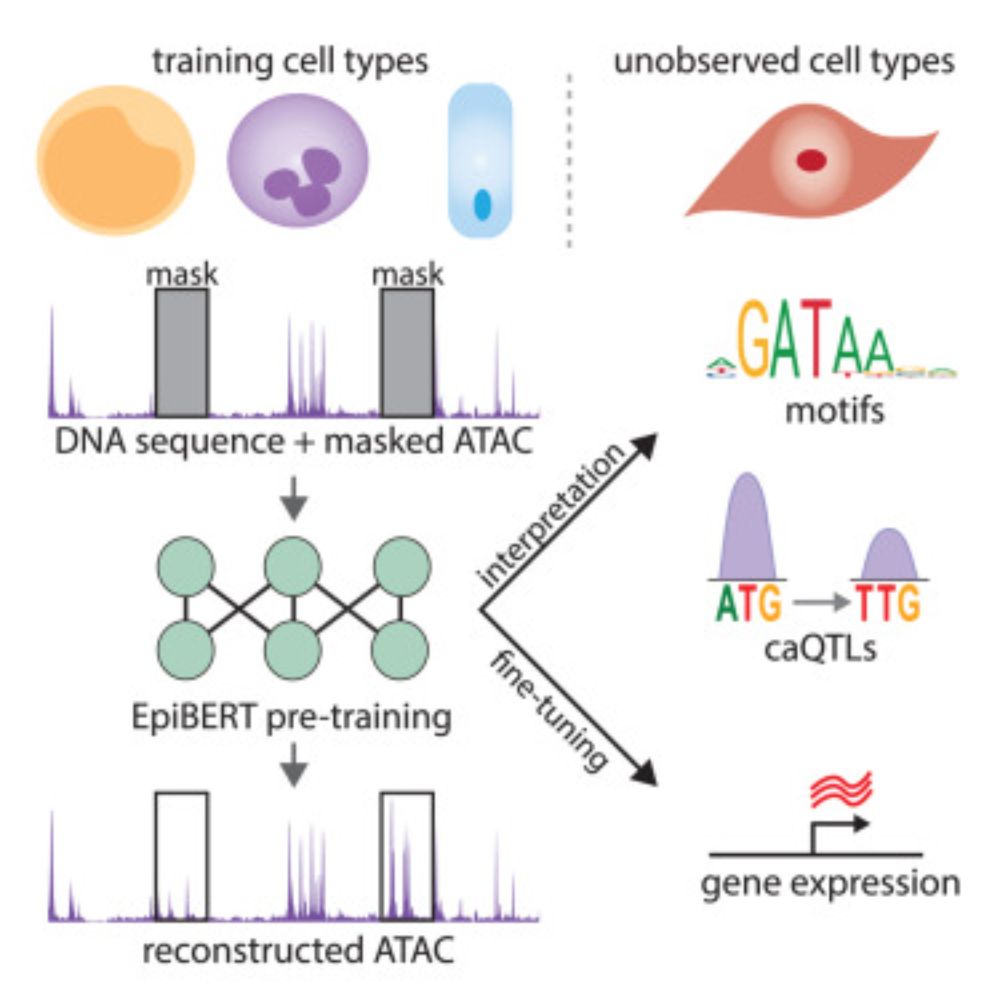
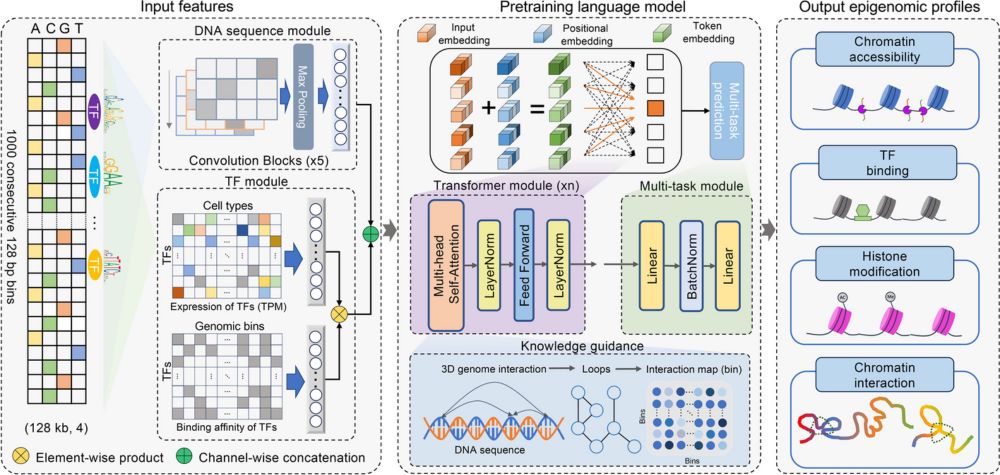
A multi-modal transformer for cell type-agnostic regulatory predictions
Javed and Weingarten et al. created a multi-modal transformer that learns generalizable
representations of genomic sequence and chromatin accessibility by utilizing a novel
masked-accessibility pre-tr...
Excited to present the results of my 20% project in collaboration with @broadinstitute.org and @danafarber.bsky.social . In our new paper we demonstrate a long-range model capable of detecting regulatory elements at distances beyond a million base pairs.
30.01.2025 16:50 — 👍 36 🔁 7 💬 1 📌 0

Topics:
Population Genomics
Evolutionary & Non-human Genomics
Cancer Genomics
Computational & Statistical Genomics
Complex Traits & Genomic Medicine
Functional Genomics
Emerging Methods & Technologies
Keynote Speakers:
Steve McCarroll, Broad Institute of MIT and Harvard
Trisha Wittkopp, University of Michigan
Discussion Leaders:
Nadav Ahituv, University of California, San Francisco
Gemma Carvill, Northwestern University
Nancy Chen, University of Rochester
Gilad Evrony, New York University
Simon Gravel, McGill University, Canada
Stephanie Hicks, Johns Hopkins University
Nada Jabado, McGill University, Canada
Andrew Kern, University of Oregon
Nicholas Mancuso, University of Southern California
Sara Mostafavi, University of Washington
Nicholas Navin, MD Anderson Cancer Center
Soumya Raychaudhuri, Broad Institute of MIT and Harvard
Gloria Sheynkman, University of Virginia School of Medicine
Peter Sudmant, University of California, Berkeley
One month to go until the CSHL Biology of Genomes abstract deadline! My all-time fav meeting, and one of the few covering genomics broadly. We have a ✨🤩 lineup of speakers, but it's the abstract talks & posters that really make the meeting - send us your best work! meetings.cshl.edu/meetings.asp...
14.01.2025 13:57 — 👍 51 🔁 21 💬 1 📌 1
a green background with red circles and a green circle in the middle
ALT: a green background with red circles and a green circle in the middle
1/2 We’re hiring a Postdoc to join us at the University of Copenhagen. Are you fascinated by 3D chromatin function and super-resolution microscopy? Then this might be the job for you. Start date 1st of April 2025, starting salary 4700 Euro/month. Apply here: candidate.hr-manager.net/ApplicationI...
29.12.2024 20:46 — 👍 29 🔁 22 💬 1 📌 0
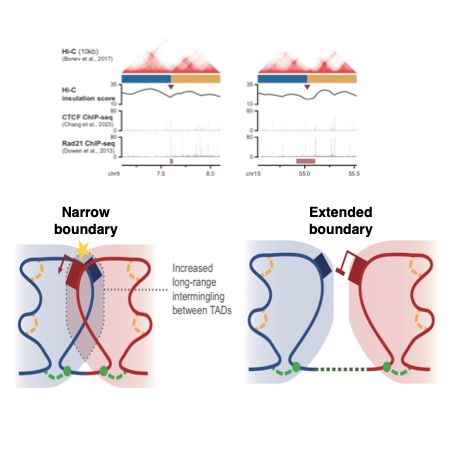
Boundaries between TADs often extend over considerable distance. But how variable is this width of boundaries? And does it matter? Find out in our new pre-print with the group of David Holcman. www.biorxiv.org/content/10.1...
26.12.2024 12:10 — 👍 37 🔁 17 💬 2 📌 1
Our ChromBPNet preprint out!
www.biorxiv.org/content/10.1...
Huge congrats to Anusri! This was quite a slog (for both of us) but we r very proud of this one! It is a long read but worth it IMHO. Methods r in the supp. materials. Bluetorial coming soon below 1/
25.12.2024 23:48 — 👍 231 🔁 89 💬 8 📌 5
Redirecting
By definition, enhancers can activate from a distance. But with increased distance between enhancer and promoter, the activation drops. To study this systematically, we build a synthetic locus: www.cell.com/molecular-ce... 1/12
02.12.2024 16:44 — 👍 226 🔁 84 💬 14 📌 11
Ever wondered how 3D chromatin rewires during differentiation? We wanted to understand this process better & distinguish if different processes or elements drive this rewiring. To do so, we used mESC=>NPC differentiation and focused on a single locus. biorxiv.org/content/10.110… 1/17
01.12.2024 14:52 — 👍 25 🔁 10 💬 3 📌 0
PhD Student, MRC Biostatistics Unit
University of Cambridge
Gates Cambridge Scholar
Bioinformatics, genetics, single-cell, statistics
Australian 🇦🇺
Human Geneticist/Genomicist working on Mendelian & rare genetic disorders to enable Precision Medicine. Opinions are my own.
@cgonzagaj everywhere 🐦🦣☁️🧵
Bioinformatics Software Engineer @ @pacbio.bsky.social 🧬🖥️. Previously at the data sciences platform @ @broadinstitute.org. Interested in high-performance software engineering, sequence alignment algorithms, and GPU programming. Views are my own.
Assistant Professor at NYU Langone. Genetics, evolution and biology of complex traits and diseases.
Statistical geneticist @ Regeneron
tweets are my own | he/him/his
Genetics, stats, complex traits. Asst. prof. at the University of Minnesota - Twin Cities. views my own. he/him - Zaidilab.org
💻 Assoc Prof of Epidemiology at JH Public Health
🛠️ RAGE Lab: Research on Ancestry in Genetic Epi
📢 loud bark, deep bite.
Scientist interested in chromatin and regulation of transcription. Klose lab, University of Oxford
Enhancers, 3D genome organisation, pluripotent stem cells
Babraham Institute and Enhanc3D Genomics
Principal AI scientist at Genentech. Previously NVIDIA, insitro, Stanford. AI + Genomics.
Group Leader
The Genome Function Laboratory
The Francis Crick Institute, London
We study transcriptional regulation and chromosome folding using an interdisciplinary approach combining wet- and dry-lab methods.
https://giorgettilab.org
@fmiscience.bsky.social
Stanford BioE, Genetics & Sarafan ChEM-H. Chan-Zuckerberg Biohub Investigator. Our lab develops and applies microfluidic assays for high-throughput biophysics and biochemistry.
Group leader at EMBL Heideberg
https://www.embl.org/groups/krebs/
Postdoctoral Fellow, Genome Biology Unit, EMBL Heidelberg
Molecular biologist with interest in transcription, chromatin, nuclear organisation, and developmental biology. Runs a lab, teaches students, and is concerned about the world we live in.
Official account of the Max Planck Society. Devoted to basic #research in #physics #astronomy #chemistry #biology #earthsciences #materialscience #mathematics #socialsciences and the #humanities; Imprint: https://www.mpg.de/imprint
Associate Professor of Applied and Computational Mathematics and Statistics and Biological Sciences at U Notre Dame. Theoretically a Neuroscientist.