
GPU-accelerated homology search with MMseqs2 - Nature Methods
Graphics processing unit-accelerated MMseqs2 offers tremendous speedups for homology retrieval from metagenomic databases, query-centered multiple sequence alignment generation for structure predictio...
MMseqs2-GPU sets new standards in single query search speed, allows near instant search of big databases, scales to multiple GPUs and is fast beyond VRAM. It enables ColabFold MSA generation in seconds and sub-second Foldseek search against AFDB50. 1/n
📄 www.nature.com/articles/s41...
💿 mmseqs.com
21.09.2025 08:06 — 👍 174 🔁 64 💬 4 📌 2
Sorry for the slow responses lots of traveling this week. We use a paired MSA for the toxin-antitoxin proteins (many rows from different species). The top row is the mutated antitoxin sequence + fixed toxin seq, and we compute the pseudolikelihood over the 4 mutated positions by masking each
10.08.2025 21:13 — 👍 1 🔁 0 💬 1 📌 0

MMseqs2 v18 is out
- SIMD FW/BW alignment (preprint soon!)
- Sub. Mat. λ calculator by Eric Dawson
- Faster ARM SW by Alexander Nesterovskiy
- MSA-Pairformer’s proximity-based pairing for multimer prediction (www.biorxiv.org/content/10.1...; avail. in ColabFold API)
💾 github.com/soedinglab/M... & 🐍
05.08.2025 08:25 — 👍 62 🔁 17 💬 0 📌 0
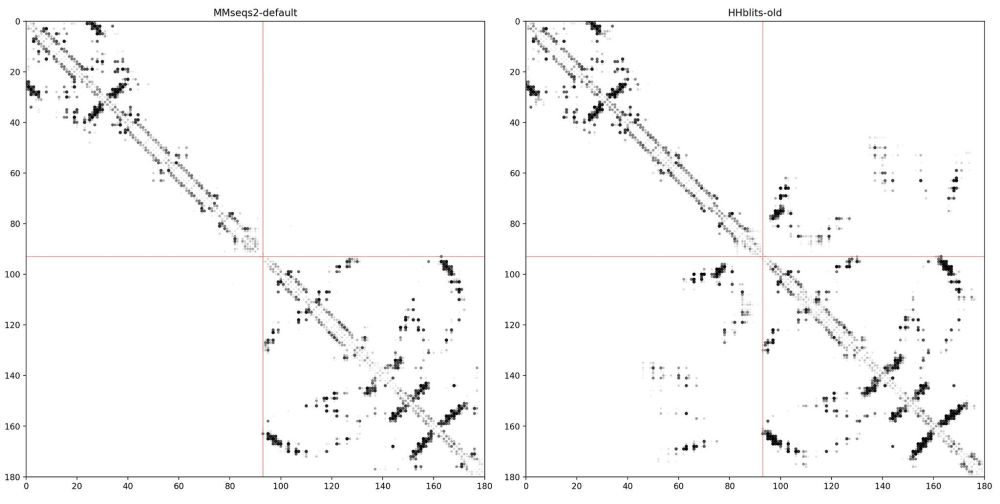
Side story: While working on the Google Colab notebook for MSA pairformer. We encountered a problem: The MMseqs2 ColabFold MSA did not show any contacts at protein interfaces, while our old HHblits alignments showed clear contacts 🫥... (2/4)
05.08.2025 07:39 — 👍 13 🔁 3 💬 1 📌 0

GitHub - yoakiyama/MSA_Pairformer
Contribute to yoakiyama/MSA_Pairformer development by creating an account on GitHub.
Our code and Google Colab notebook can be found here
github.com/yoakiyama/MS...
colab.research.google.com/github/yoaki...
Please reach out with any comments, questions or concerns! We really appreciate all of the feedback from the community and are excited to see how y'all will use MSA Pairformer :)
05.08.2025 06:29 — 👍 4 🔁 0 💬 0 📌 0

Special thanks to all members of our team! Their mentorship and support are truly world-class.
And a huge shoutout to the entire solab! I'm so grateful to work with these brilliant and supportive scientists every day. Keep an eye out for exciting work coming out from the team!
05.08.2025 06:29 — 👍 2 🔁 0 💬 1 📌 0

Thanks for tuning in--we've already received incredibly valuable feedback from the community and will continue to update our work!
We're excited for all of MSA Pairformer's potential applications for biological discovery and for the future of memory and parameter efficient pLMs
05.08.2025 06:29 — 👍 7 🔁 1 💬 1 📌 0

We made some updates to MSA pairing with MMseqs2 for modeling protein-protein interactions! Mispairing sequences leads to contamination of non-interacting paralogs. We use genomic proximity to improve pairing, and find that MSA Pairformer's predictions reflect pairing quality
05.08.2025 06:29 — 👍 1 🔁 0 💬 2 📌 0

We also looked into how perturbing MSAs effects contact prediction. Interestingly, unlike MSA Transformer, MSA Pairformer doesn't hallucinate contacts after ablating covariance from the MSA. Hints at fundamental differences in how they extract pairwise relationships
05.08.2025 06:29 — 👍 0 🔁 0 💬 1 📌 0

We ablate triangle updates and replace it with a pair updates analog. As expected, contact precision deteriorates, and the false positives are enriched in indirect correlations. These results suggest the role of triangle updates in disentangling direct and indirect correlations
05.08.2025 06:29 — 👍 0 🔁 0 💬 1 📌 0

Whereas the ESM2 family models show an interesting trade-off between contact precision and zero-shot variant effect prediction, MSA Pairformer performs strongly in both
P.S. this figure slightly differs from what's in the preprint and will be updated in v2 of the paper!
05.08.2025 06:29 — 👍 0 🔁 0 💬 1 📌 0

Using a library of mutants at four key ParD3-ParE3 toxin-antitoxin interface residues from Aakre et al. (2015), we find that MSA Pairformer's pseudolikelihood scores better discriminate binders and non-binders, directly related to its ability to model the interaction
05.08.2025 06:29 — 👍 1 🔁 0 💬 2 📌 0
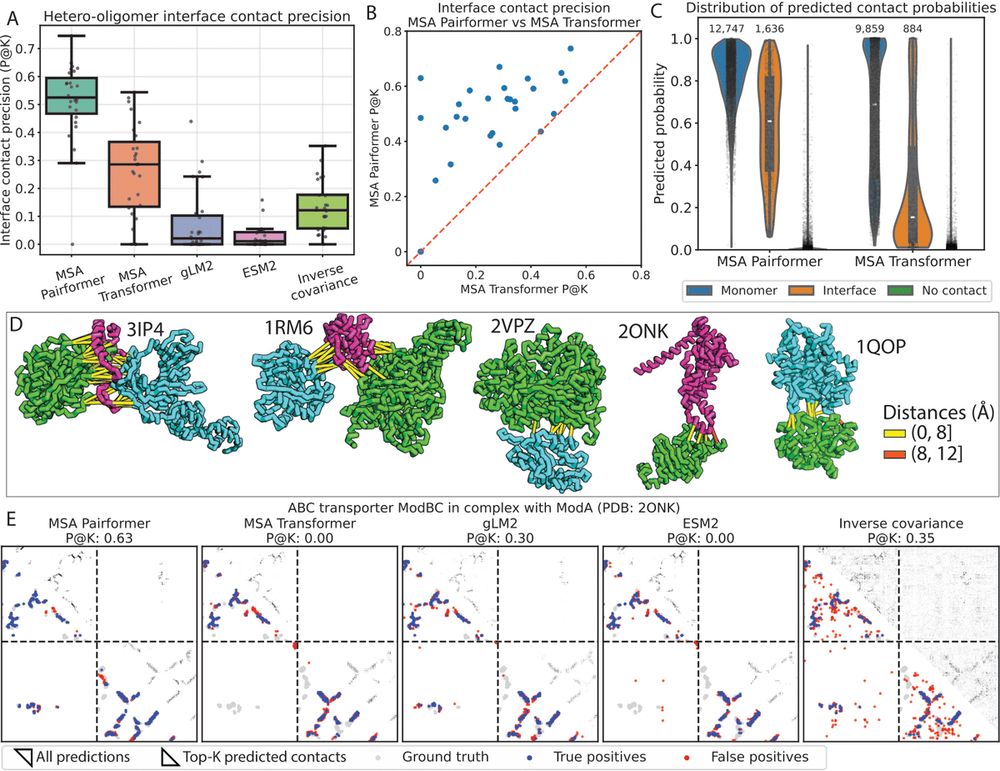
Beyond monomeric structures, accurate prediction of protein-protein interactions is crucial for understanding protein function. MSA Pairformer substantially outperforms all other methods in predicting residue-residue interactions at hetero-oligomeric interfaces
05.08.2025 06:29 — 👍 14 🔁 4 💬 1 📌 0

On unsupervised long-range contact prediction, it outperforms MSA Transformer and all ESM2 family models, suggesting that its representations more accurately capture structural signals from evolutionary context
05.08.2025 06:29 — 👍 0 🔁 0 💬 1 📌 0

We introduce MSA Pairformer, a 111M parameter memory-efficient MSA-based protein language model that builds on AlphaFold3's MSA module to extract evolutionary signals most relevant to the query sequence via a query-biased outer product
05.08.2025 06:29 — 👍 1 🔁 0 💬 1 📌 0
Current efforts to improve self-supervised protein language modeling focus on scaling model and training data size, requiring vast resources and limiting accessibility. Can we
1) scale down protein language modeling?
2) expand its scope?
05.08.2025 06:29 — 👍 1 🔁 0 💬 1 📌 0

Scaling down protein language modeling with MSA Pairformer
Recent efforts in protein language modeling have focused on scaling single-sequence models and their training data, requiring vast compute resources that limit accessibility. Although models that use ...
Excited to share work with
Zhidian Zhang, @milot.bsky.social, @martinsteinegger.bsky.social, and @sokrypton.org
biorxiv.org/content/10.1...
TLDR: We introduce MSA Pairformer, a 111M parameter protein language model that challenges the scaling paradigm in self-supervised protein language modeling🧵
05.08.2025 06:29 — 👍 95 🔁 43 💬 1 📌 1
Images, AI, ML, and ALife.
Former: @hampshirecolg
@brandeisuniversity.bsky.social
@harvardmed.bsky.social
@uidaho.bsky.social
@hhmijanelia.bsky.social
@mdc-berlin.bsky.social
@ORNL.
Now @chanzuckerberg.bsky.social.
Opinions are mine.
Postdoc @ ETH Zurich / SIB Swiss Institute of Bioinformatics | Protein bioinformatics
Computational genomics and human genetics. Associate Professor @ Columbia University
Lab: http://www.columbia.edu/~ys2411/
🔬 Assistant Prof, Pathology @Duke | Director, Clin Micro Lab
🧫 Former Clin Micro Fellow @Memorial Sloan Kettering
👩🏻🔬 Former Postdoc @broadinstitute.org
🎓 PhD @The Rockefeller University
Focus: Diagnostics, AMR, Structural Biology
assistant professor @mskcancercenter.bsky.social
clareaulab.com
A scientific journal publishing cutting-edge methods, tools, analyses, resources, reviews, news and commentary, supporting life sciences research. Posts by the editors.
Chief Editor of Nature Biomedical Engineering. Ph.D. Previously at Nature Methods. @rita_strack on the place formerly known as twitter.
Principal Bionformatician
@nanopore. Ex: Postdoctoral fellow @ NIH; Researcher @ CAB. Views are my own; #StandWithUkraine Support Ukraine!
Senior AI Scientist at Absci (@abscibio)
Asst prof at HMS, PI at DFCI
Designing proteins
polizzilab.org
Postdoc @ Debbie Marks Lab, Harvard | Prev. PhD @ MIT EECS || ML for Proteins + Viruses 🦠
Associate Professor, MIT
Still thinking about the 10^9 mutations generated in your microbiome today.
Website: http://lieberman.science
Professor & Scientist @uspoficial.bsky.social 🇧🇷. Visiting @mit.edu. Scholar @ieausp.bsky.social. Computational (bio)chemistry and biophysics.
Web: http://gaznevada.iq.usp.br
🐦 : x.com/ArantesLab
🎵 : www.dinamicas.art.br
Teaching Biology at NCSSM
Discover the Languages of Biology
Build computational models to (help) solve biology? Join us! https://www.deboramarkslab.com
DM or mail me!
PhD at EPFL | Biophysics and Machine Learning
PhD student @sangerinstitute
Research in AI for Protein Design @Harvard | Prev. CS PhD @UniofOxford, Maths & Physics @Polytechnique














