It's publication day, and Cold Spring Harbor Laboratory Press is running a promotion bundling the hard cover and e-book, and other offers:
cshlpress.com/default.tpl?...
#EpigeneticsBook
21.10.2025 15:18 — 👍 12 🔁 6 💬 1 📌 0
A challenge faced by many families undergoing genetic testing is that it's not feasible to collect samples from both biological parents. Hoping duoNovo can help address this.
11.10.2025 19:35 — 👍 0 🔁 0 💬 0 📌 0
Thanks, John. Hope this ends up being useful!
11.10.2025 19:20 — 👍 2 🔁 0 💬 0 📌 0
YouTube video by InfiniteHistoryProject MIT
David Baltimore
One of the greats of 20th century biology. What a life.
www.youtube.com/watch?v=v9Em...
08.09.2025 18:21 — 👍 0 🔁 0 💬 0 📌 0
yeah it's a very vanilla setting
08.08.2025 03:14 — 👍 0 🔁 0 💬 0 📌 0
That's amusing for sure. But still, the approach is imo quite interesting and there's interesting results, e.g. the lower MAE for dispersion.
08.08.2025 01:21 — 👍 0 🔁 0 💬 1 📌 0
My book Epigenetics: History, Molecules and Diseases will be published in exactly one month (September 2).
Finalizing cover (looks great), probably going to printer this week.
Nervous anticipation is the mood right now.
02.08.2025 20:48 — 👍 76 🔁 9 💬 6 📌 1


#apaperaday is back from holiday. Yuzu is still with the birbsitter so a holiday picture it is. Today's pick is from @nejm.org by Musunuru et al on an N=1 case (patient with severe metabolic disease) treated by base editing (CRISPR therapy). DOI: 10.1056/NEJMoa2504747 Very good way to restart!
27.05.2025 04:35 — 👍 8 🔁 3 💬 1 📌 2
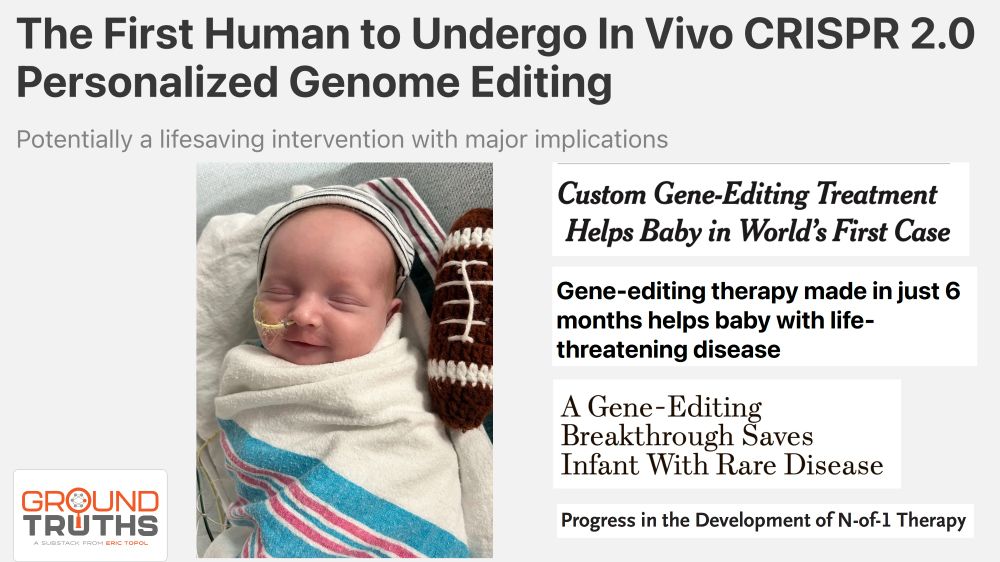
Multiple headlines about the first CRISPR 2.0 personalized genome editing and photo of the baby treated
This week's genome editing triumph is a big deal.
Here's why
erictopol.substack.com/p/the-first-...
18.05.2025 14:37 — 👍 625 🔁 136 💬 23 📌 19
Reminded of Ed Yong’s response to this question:
“If I cover a preprint, I talk to 2-3 experts to get their views before writing the story.
That’s exactly what I do if I cover a journal article…”
18.05.2025 12:59 — 👍 96 🔁 31 💬 6 📌 1
Looking forward to this next week
16.05.2025 09:46 — 👍 2 🔁 0 💬 0 📌 0
Beautiful combination of population-scale genetic analyses and mouse work. Damaging variants in a highly constrained chromatin remodeler (CHD1) are better tolerated in males, with mouse experiments indicating a protective effect of androgens.
09.05.2025 01:37 — 👍 0 🔁 0 💬 0 📌 0

Design principles of cell-state-specific enhancers in hematopoiesis
Screen of minimalistic enhancers in blood progenitor cells demonstrates widespread
dual activator-repressor function of transcription factors (TFs) and enables the model-guided
design of cell-state-sp...
Out in Cell @cp-cell.bsky.social: Design principles of cell-state-specific enhancers in hematopoiesis
🧬🩸 screen of fully synthetic enhancers in blood progenitors
🤖 AI that creates new cell state specific enhancers
🔍 negative synergies between TFs lead to specificity!
www.cell.com/cell/fulltex...
🧵
08.05.2025 16:06 — 👍 139 🔁 58 💬 4 📌 8
+1 Anshul
Part of this means reviewers not ripping on manuscripts that explicitly mention limitations *for those limitations*
08.04.2025 13:22 — 👍 24 🔁 4 💬 0 📌 0
Just a gentle reminder that deceptive hyping in scientific publications (which includes preprints) is actually antithetical to the core mission of the scientific process. We can stay grounded, truthful, humble while being ambitious. Revealing caveats, pitfalls & limitations speeds up progress.
08.04.2025 09:04 — 👍 95 🔁 16 💬 3 📌 0
Great new review @natrevgenet.bsky.social by @sedlazeck.bsky.social @timp0.bsky.social and @yileifu.bsky.social! 👇highlighting both the promise of long read DNA methylation analysis and remaining challenges such as the difficulty to benchmark 5hmC and non-CG 5mC given their low abundance👏
30.03.2025 20:56 — 👍 14 🔁 6 💬 0 📌 1
Cohesin as an essential disruptor of chromosome organization
Cohesin is a multi-subunit molecular machine that is able to create lateral chromatin loops within a linear chromosome fiber. Despite intense study, a…
Plenty of work required to rigorously test the proposed hypothesis but my initial thoughts are this perspective by Adrian Bird in @cp-molcell.bsky.social may well be the most significant conceptual advance in the 3D genome organisation field in years:
www.sciencedirect.com/science/arti...
21.03.2025 23:19 — 👍 29 🔁 9 💬 3 📌 2
I just the word epigenetics; it has so many meanings that it's next to useless. I prefer less ambigous terms: Epigenomic data for chromatin-related data (including methylation); gene regulatory mechanisms for the general phenomenon of how genes are regulated, etc.
19.03.2025 10:29 — 👍 7 🔁 4 💬 3 📌 0
The analogy of omics data normalization and cooking:
some processing is usually necessary, overcooking makes it bland, highly over-processed foods are unhealthy, and the quality of the ingredients matters.
27.02.2025 08:01 — 👍 76 🔁 23 💬 1 📌 5
Important to mention that duoNovo was developed and tested using sequencing data from the NIH-funded @gregor-research.bsky.social consortium
Feedback and comments are especially welcome - as I mentioned earlier, we really hope that duoNovo can be broadly useful (n/n)
27.02.2025 23:07 — 👍 1 🔁 0 💬 0 📌 0
In the preprint we show that this simple approach performs very well in practice. Among rare variants - which are of course the most likely to be disease-causing - duoNovo has perfect accuracy in our evaluations (7/n)
27.02.2025 23:04 — 👍 0 🔁 0 💬 1 📌 0
Then, using just one parent, we can tell whether the haplotype containing a given variant of interest was inherited from the available parent. If that's the case, then the variant must be de novo - we don't need to look at the other parent. (6/n)
27.02.2025 23:02 — 👍 0 🔁 0 💬 1 📌 0
Our method solves this problem using a simple idea. We first reconstruct haplotypes using read-backed phasing enabled by long read sequencing. This allows us to phase variants that we want to test for de novo status (which isn't possible with non-read-backed phasing approaches). (5/n)
27.02.2025 23:01 — 👍 0 🔁 0 💬 1 📌 0
Which is why de novo status is used as a strong criterion of variant pathogenicity.
However, to identify a variant as de novo, both biological parents have to be sequenced. For millions of families, this is impossible for a variety of reasons. (4/n)
27.02.2025 23:00 — 👍 0 🔁 0 💬 1 📌 0
Genomics enthusiast | work at PacBio | running | coffee | cycling | cake | gigs | all my thoughts and opinions are my own
🧬Founded in 1948, the American Society of Human Genetics (ASHG) is the primary professional membership organization for #humangenetics specialists worldwide.
www.ashg.org
Staff scientist at @PacBio; formerly @hudsonalpha; avid gamer; opinions are my own
AI slop (except one) enjoyer
Research Scientist at Stanford University. Interested in everything genomics and computational biology.
Our laboratory seeks to understand how chromosome structure relates to genome functions
Medicinal chemist / chemical biologist, author of “In the Pipeline” at http://science.org/blogs/pipeline. derekb.lowe@gmail.com and on Signal at Dblowe.18
All opinions are mine; I don’t speak for my employer in any way.
Science. Biology. Progress.
Founding Editor of Asimov Press. Subscribe at press.asimov.com!
Postdoc at MD Anderson: Omics of reproduction, development, cancer, and conflict | PhD, Penn State: functional genomics in social insects
Opinions are my own.
https://seantbresnahan.com
https://bhattacharya-lab.com
Cell Bio@Harvard Med- dedicated to unraveling how the machines of the cell work. Specializing in membrane/organelles, ubiquitin & protein quality control, chromatin regulation, proteomics, metabolism, and sensory perception.https://cellbio.hms.harvard.edu/
Studying cellular #stress responses, particularly #senescence and its impact on #immune response, #ageing and #cancer, #tumorigenesis at Cancer Research UK CI @cruk-ci.bsky.social, University of Cambridge @cam.ac.uk
Website: naritalab.com
UK Doctor specialising in Clinical Genetics and Genomics. Researcher investigating Wiedemann-Steiner syndrome, chromatin disorders and genetics of orofacial clefting. All views are my own.
🐏 phd’ing in bioinformatics & computational biology @ unc chapel hill | phanstiel lab | interests: genome org & function 🧬 | hhmi gilliam '24 | nsf grfp ‘22 | 🐅 occidental college ‘21 |🎙host, from where does it stem? | 🏘️ co-founder @sciforgood.bsky.social
Science Communicator in Genomics, Lead Public Affairs at NHGRI, lover of pop culture and romcoms
ICREA Research Professor. Evolution, Genetics, Neuroscience, Linguistic Cognition
Principal Researcher in BioML at Microsoft Research. He/him/他. 🇹🇼 yangkky.github.io
Associate Professor at Johns Hopkins studying human evolutionary and reproductive genetics https://mccoy-lab.org










