
Check out Dr. Mendez-Dorantes' recent preprint for more details on his exciting research! www.biorxiv.org/content/10.1... 42/
09.05.2025 14:41 — 👍 4 🔁 3 💬 1 📌 0@carlosmendezphd.bsky.social
Cientifico https://scholar.google.com/citations?user=9UIDNdwAAAAJ&hl=en

Check out Dr. Mendez-Dorantes' recent preprint for more details on his exciting research! www.biorxiv.org/content/10.1... 42/
09.05.2025 14:41 — 👍 4 🔁 3 💬 1 📌 0Dr. Carlos Mendez-Dorantes @carlosmendezphd.bsky.social from Dr. Kathleen Burns' lab @danafarber.bsky.social is telling us about the LINE-1 retrotransposon and its causal role in genome instability and chromosomal rearrangements in a wide variety of cancers. 41/
09.05.2025 14:41 — 👍 3 🔁 2 💬 1 📌 0
It is a cloudy day in Chicago, but the science is shining here inside the Gleacher Center at the University of Chicago at the 2025 JCC Annual Symposium.
First up this morning: science talks from our 3rd year JCC Fellows ... buckle up!
1 / a lot
LINE-1 retrotransposons drive genomic instability and immune activation in cancer. In this review, Mendez-Dorantes and Burns discuss their molecular mechanisms, genomic consequences, and diagnostic and therapeutic targetability.
Read more here:
➡️ tinyurl.com/genesdev351051
Congrats @carlosmendezphd.bsky.social!!!
03.01.2025 14:28 — 👍 1 🔁 1 💬 0 📌 0
Chromosomal rearrangements and instability caused by the LINE-1 retrotransposon
www.biorxiv.org/content/10.1...
Chromosomal rearrangements and instability caused by the LINE-1 retrotransposon https://www.biorxiv.org/content/10.1101/2024.12.14.628481v1
18.12.2024 07:37 — 👍 2 🔁 2 💬 0 📌 0Chromosomal rearrangements and instability caused by the LINE-1 retrotransposon https://www.biorxiv.org/content/10.1101/2024.12.14.628481v1
18.12.2024 07:37 — 👍 8 🔁 5 💬 0 📌 1
Also, check out a pre-print from the lab of Jose Tubio: they analyzed cancer genomes with a high burden of somatic L1 insertions using long-read WGS. They also uncovered that synchronous L1 lesions contribute to chromosomal rearrangements.
www.biorxiv.org/content/10.1...
Huge thanks to our funding sources, including to
@jcchildsfund.bsky.social for supporting my postdoctoral work.
Shout out to my fellow co-first authors Xi Zeng and Jennifer Karlow and co-authors Phil, Serafina and Jupiter, our genomics experts and collaborators Eunjung A Lee and Cheng-Zhong Zhang and my fantastic mentor Kathleen Burns.
19.12.2024 21:53 — 👍 1 🔁 0 💬 1 📌 0
Check out our manuscript for many more examples, our insertional mutagenesis analysis, and our evidence showing L1 can cause foldback rearrangements and contribute to chromothriptic chromosomes.
www.biorxiv.org/content/10.1...
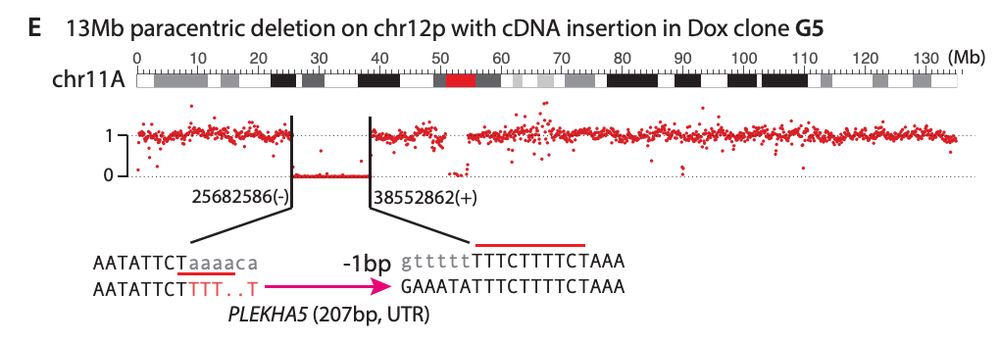
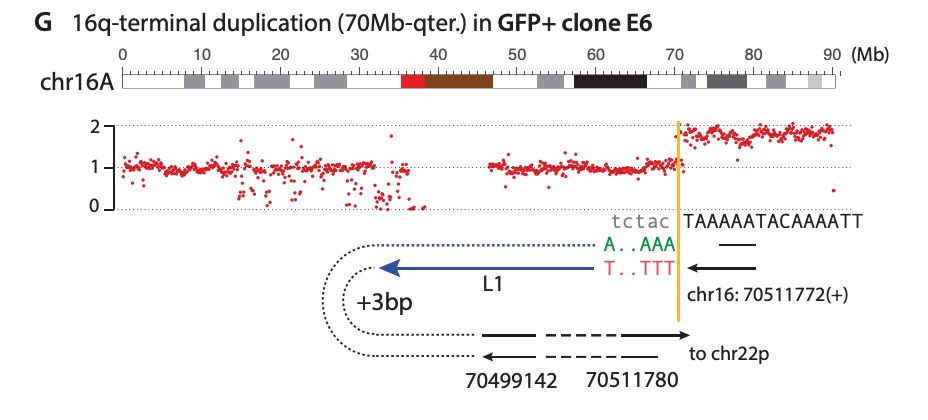
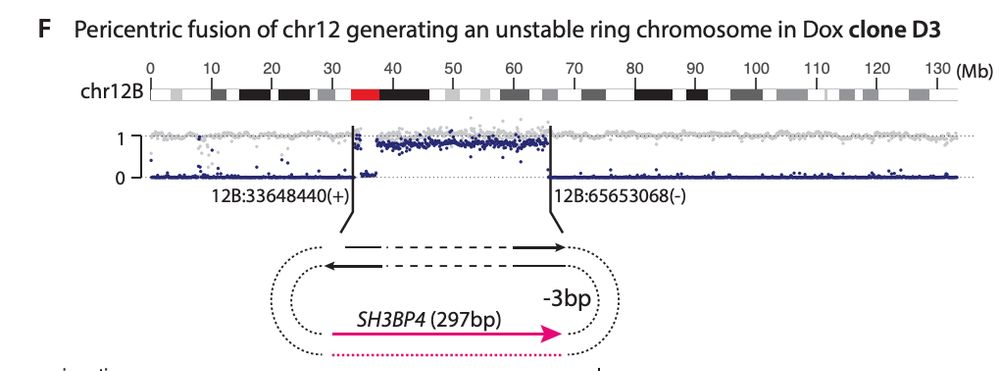
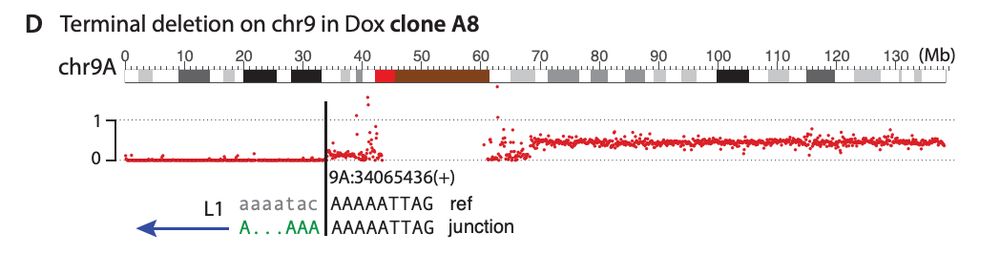
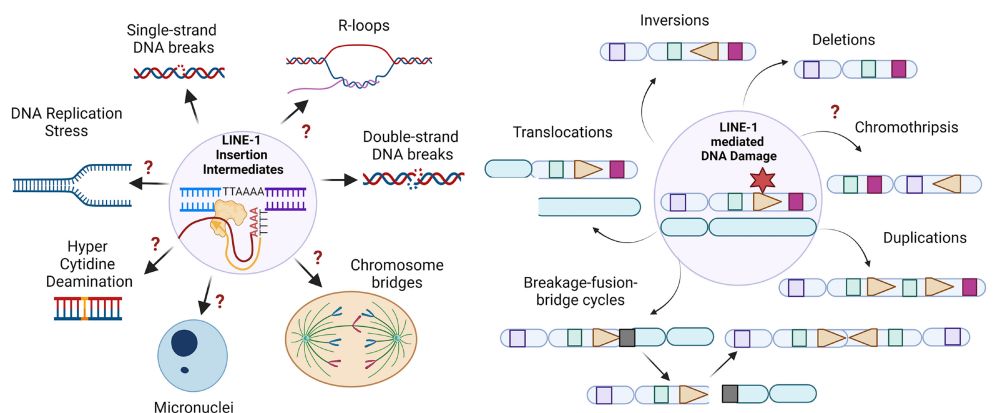
We also find L1-mediated DSBs can generate diverse types of large-scale intrachromosomal rearrangements, including deletions and duplications. Interestingly, we find L1 caused a pericentric ring chromosome!
19.12.2024 21:52 — 👍 1 🔁 0 💬 1 📌 0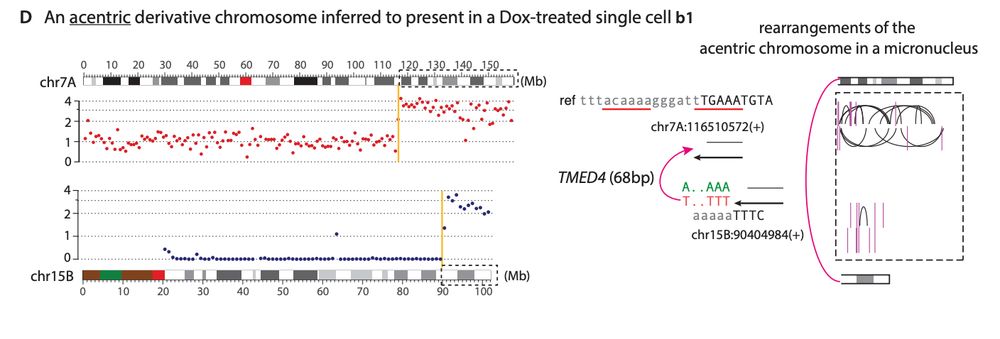
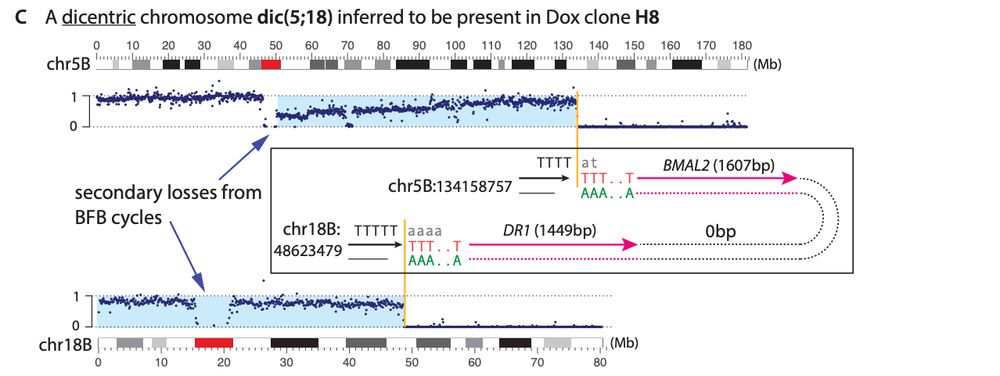
Similar to balanced translocations, we find that end joining between distal L1-mediated DSBs can generate dicentric or acentric chromosomes that result in complex rearrangements via breakage-fusion-bridge cycles or DNA fragmentation (chromothripsis).
19.12.2024 21:51 — 👍 1 🔁 0 💬 1 📌 0The latter suggest that contribution of L1 retrotransposition to chromosomal rearrangements based on canonical L1 genomic footprints may be underestimated in our current cancer genome analyses.
19.12.2024 21:51 — 👍 1 🔁 0 💬 1 📌 0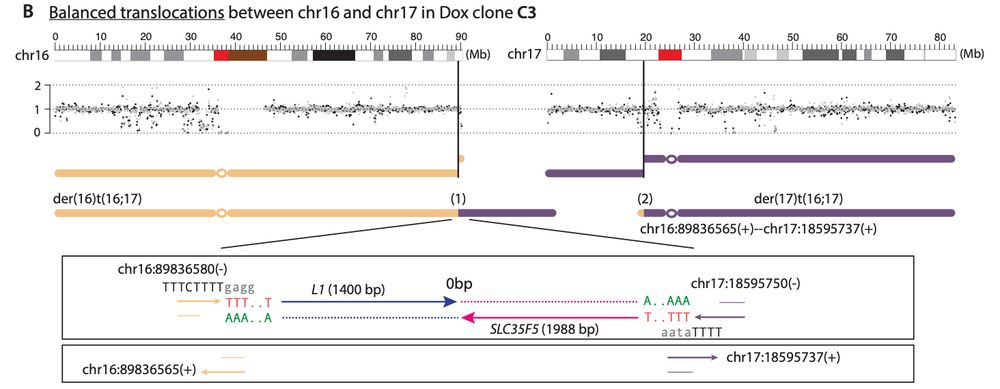
For example, we show that L1-mediated DSBs from two distinct chromosomes can generate reciprocal balanced translocations (DNA copy number neutral), with one of the translocation junctions showing clear L1 TPRT evidence and the other lacking obvious evidence of L1 TPRT.
19.12.2024 21:50 — 👍 1 🔁 0 💬 1 📌 0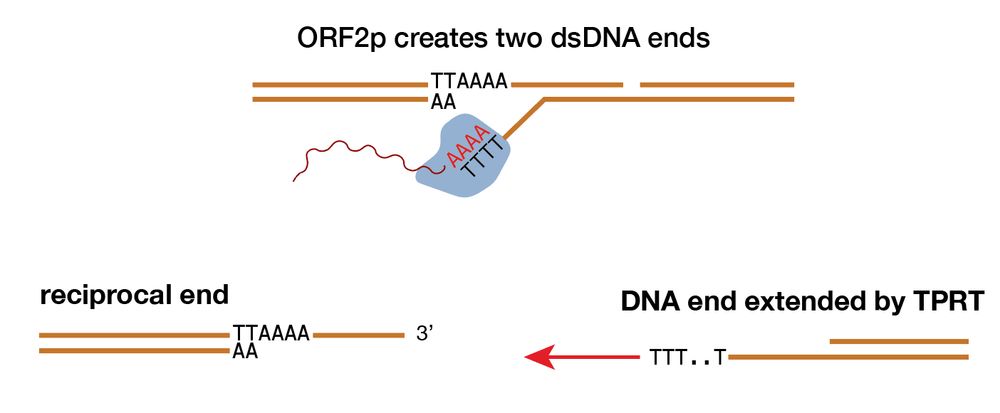
Here, we highlight the recombinogenic potential of DNA break ends of L1-mediated DSBs, with the DNA ends extended by TPRT leading to rearrangement junctions with L1 insertions and the reciprocal DNA ends leading to rearrangement junctions without evidence of L1 TPRT.
19.12.2024 21:50 — 👍 1 🔁 0 💬 1 📌 0Mechanistically, we found these alterations arise from DNA double-strand breaks (DSBs) generated by L1 encoded endonuclease and reverse transcriptase ORF2p, suggesting chromosomal breakage is a step in retrotransposition, or a commonly occurring risk of retrotransposition.
19.12.2024 21:49 — 👍 1 🔁 0 💬 1 📌 0After expression of L1 in nearly diploid cells, we found L1 causes both local and long-range chromosomal rearrangements, small and large segmental copy-number alterations, and subclonal copy-number heterogeneity due to ongoing chromosomal instability.
19.12.2024 21:49 — 👍 1 🔁 0 💬 1 📌 0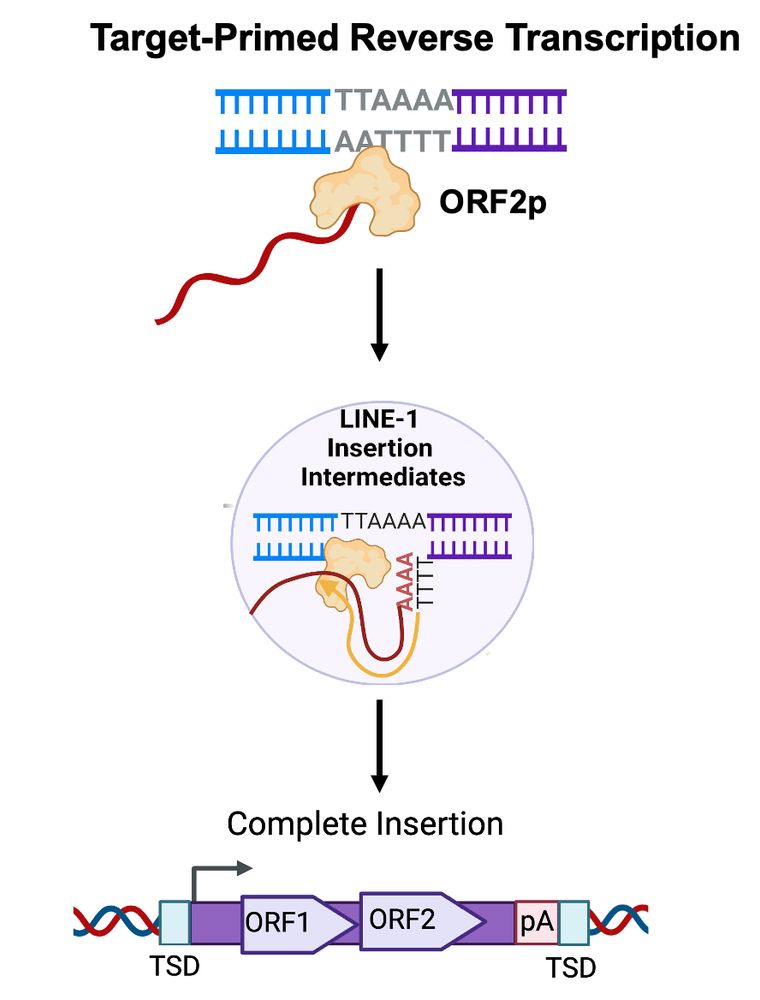
L1 retrotransposition occurs via target-primed reverse transcription (TPRT): L1 ORF2p nicks genomic DNA (3′-AA/TTTT-5′) to form a primer-template structure and then reverse transcribes the RNA to create L1 cDNA that is converted to double-stranded DNA as an insertion
19.12.2024 21:48 — 👍 1 🔁 0 💬 1 📌 0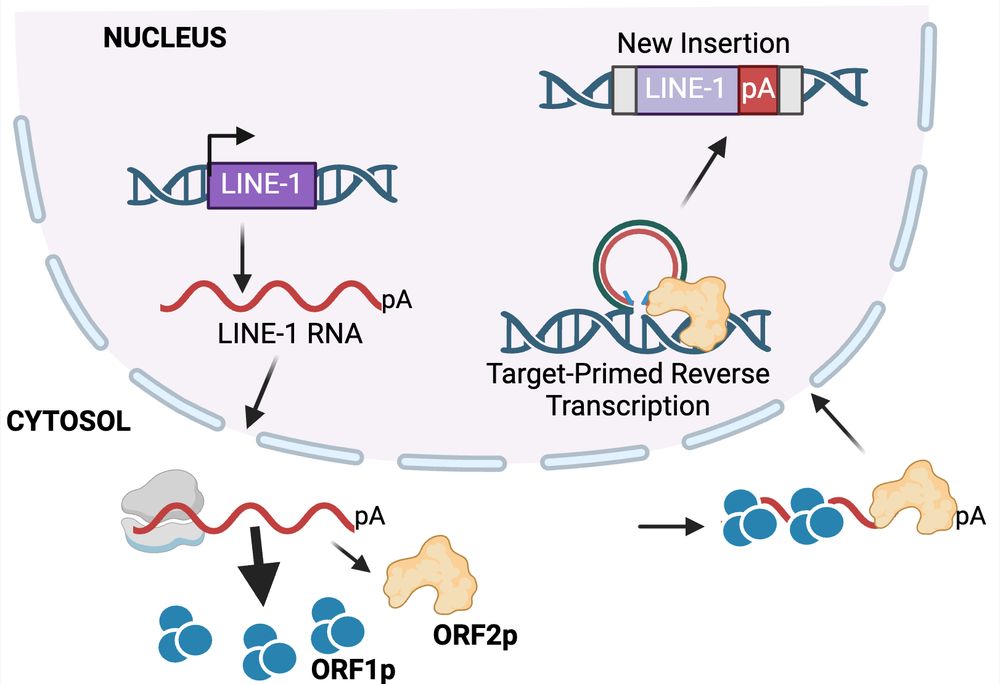
L1 is the only active protein-coding transposon in humans: ORF1p (RNA binding protein) and ORF2p (endonuclease/reverse transcriptase).
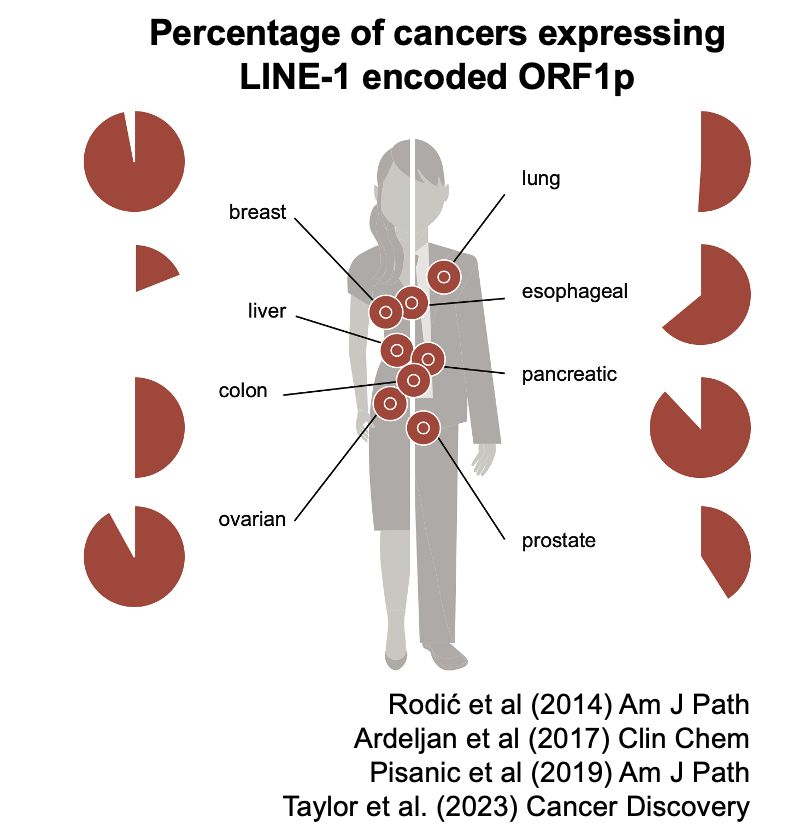
My mentor Dr. Burns established L1 is derepressed in cancers, showing ORF1p detection in malignant tissues and somatic L1 copies in cancer genomes

Check out our latest pre-print: we show that the LINE-1 retrotransposon is a potent source of chromosomal rearrangements and instability in addition to insertional mutagenesis using shotgun and long-read WGS.
www.biorxiv.org/content/10.1...