Ready to TANGO? 💃 In our new publication, Beata Turoňová & Markus Schreiber (@ms314.bsky.social, @becklab.bsky.social) present TANGO: the first cryoET framework to analyze the relation between particle positions and orientations 🪩 All in a single user-friendly package! #CryoET #OpenScience
12.02.2026 13:21 — 👍 30 🔁 10 💬 1 📌 2
We are excited to share our new preprint which is now available to read on biorXiv: doi.org/10.64898/202... 🎉🎉🎉
13.01.2026 14:38 — 👍 116 🔁 36 💬 2 📌 6
#teamtomo strikes again! Check out our new preprint on the enigmatic vault and its newly discovered association with membranes in situ.
16.12.2025 09:42 — 👍 17 🔁 1 💬 0 📌 0
That is of course what I tried... With both 3D subtomos from Warp1 (Desktop) and Warp2 (unix)
12.12.2025 17:16 — 👍 0 🔁 0 💬 1 📌 0
So, for my test case the pseudosubtomos (3D option in Relion5) already provided a better map than the 2D stacks but still not as detailed and complete as warp 3D subtomos in Relion3.
12.12.2025 15:38 — 👍 2 🔁 0 💬 0 📌 0
Can you use 3D subtomos in Relion5? I recall a message from Sjors at some point that this is no longer supported which matches my experience (from maybe 6 months ago).
12.12.2025 11:44 — 👍 1 🔁 0 💬 1 📌 0
I currently still use 3D warp subtomos in Relion3 because either version of 2D input data (pseudosubtomos or 2D stacks) give considerably worse results in Relion5 (tested for small particles).
12.12.2025 11:43 — 👍 1 🔁 1 💬 0 📌 0
Exciting new paper from my colleague @deglushk.bsky.social in the @becklab.bsky.social! It introduces a fast, robust tool for measuring membrane thickness in tomograms #TeamTomo.
06.11.2025 08:37 — 👍 8 🔁 0 💬 0 📌 0

Orange and green poster of the In-situ structural biology by Cryo-FIB and Cryo-ET in Frankfurt am Main, with EMBO logo. The poster promotes childcare grants, travel grants and that this is a sustainable event.
Have you registered to the next EMBO Practical Course on In-situ structural biology by Cryo-FIB and Cryo-ET? 🧊 Join us in Frankfurt for hands-on training on cryo-techniques and networking with experts!
🔗 Apply by Dec 8, 2025: meetings.embo.org/event/26-in-...
📍 Frankfurt | 📆 April 12–20, 2026
23.10.2025 09:10 — 👍 14 🔁 9 💬 0 📌 0

Icecream: High-Fidelity Equivariant Cryo-Electron Tomography
Cryo-electron tomography (cryo-ET) visualizes 3D cellular architecture in near-native states. Recent deep-learning methods (CryoCARE, IsoNet, DeepDeWedge, CryoLithe) improve denoising and artifact cor...
Our colleagues Vinith Kishore and Valentin Debarnot from the @ivandokmanic.bsky.social lab have come up with an amazing deep learning tool for denoising and filling the missing wedge in #cryoET data. I'm pleased to introduce Icecream🍧
23.10.2025 11:12 — 👍 53 🔁 20 💬 3 📌 5
Congrats, this is really cool! I already tried it on one of my favorite tomos and the result looks fantastic.
23.10.2025 11:29 — 👍 2 🔁 0 💬 1 📌 0

Figure 1 from the preprint
Check out our preprint! With new molecular mechanisms, 140 subtomogram averages, and ~600 annotated cells under different conditions, we @embl.org were able to describe bacterial populations with in-cell #cryoET. And there’s a surprise at the end 🕵️
www.biorxiv.org/content/10.1...
#teamtomo
15.10.2025 06:26 — 👍 114 🔁 35 💬 8 📌 4
Great work! So cool to see how far the field has already advanced in just a few years.
15.10.2025 06:51 — 👍 0 🔁 0 💬 1 📌 0

The software acts as a computational shortcut to denser sampling, which is often not feasible due to radiation damage limits. → Better angular sampling, sharper segmentation, improved particle localization.
10.10.2025 10:35 — 👍 2 🔁 0 💬 0 📌 0

Optimal tilt-increment for cryo-ET
Cryo-electron tomography (cryo-ET) enables high-resolution, three-dimensional imaging of cellular structures in their native, frozen state. However, image quality is limited by a trade-off between ang...
More great cryo-ET benchmarking work from my colleagues Maarten Tuijtel, Tomáš Majtner, Beata Turoňová, & @becklab.bsky.social: Systematic study of how tilt increment choice affects downstream TM & STA. Preprint has the optimally balanced tilt increment. #TeamTomo www.biorxiv.org/content/10.1...
23.08.2025 09:18 — 👍 32 🔁 7 💬 0 📌 2

We’re hosting the @embo.org Practical Course on In-situ structural biology by CryoFIB and CryoET in April 2026! 🧬❄️ You can expect hands-on training, cutting-edge methods, and a chance to work alongside leading experts in structural biology. 🔗 Apply by Dec 8, 2025: meetings.embo.org/event/26-in-...
18.08.2025 11:29 — 👍 32 🔁 18 💬 0 📌 2
Welcome to TomoGuide
A step-by-step Cryo-ET guide
Hey #TeamTomo,
Ever been in need of a tutorial about the fundamentals of cryo-electron tomography? From preprocessing raw frames to high-res subtomogram averaging?
That's why @florentwaltz.bsky.social and I made this website!
tomoguide.github.io
Follow the thread 1 /🧵
#CryoET #CryoEM 🔬🧪
06.05.2025 16:26 — 👍 116 🔁 41 💬 5 📌 7
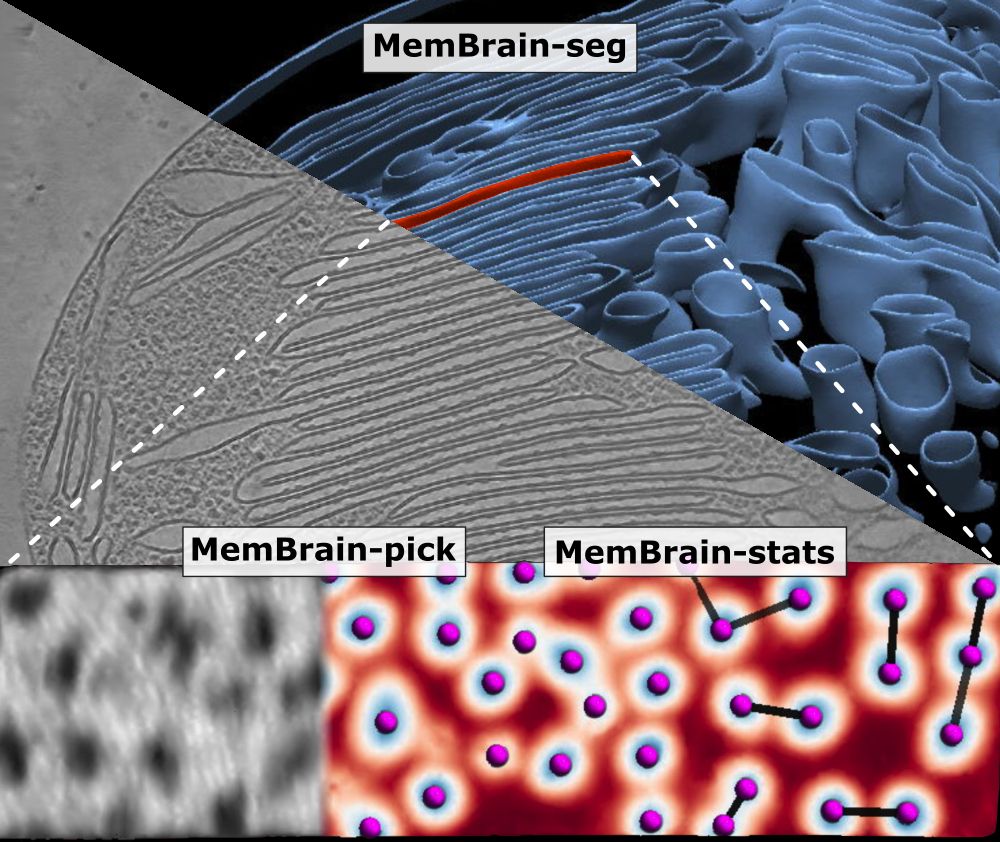
🦠🧠 MemBrain update! 🧠🦠
We’ve updated our preprint! It now covers the full MemBrain v2 pipeline for end-to-end membrane analysis in #CryoET: segmentation, particle picking, and spatial statistics.
🔗 Preprint: doi.org/10.1101/2024...
🔗 Code: github.com/CellArchLab/...
🧵(1/6) #TeamTomo
25.04.2025 07:28 — 👍 79 🔁 34 💬 2 📌 6
Looking for a tool to measure membrane thicknesses in tomograms (#TeamTomo)? Or curious about thickness differences within and across organelles? Check out my fellow @becklab.bsky.social PhD student @deglushk.bsky.social's excellent preprint:
www.biorxiv.org/content/10.1...
12.04.2025 19:29 — 👍 17 🔁 4 💬 0 📌 0
Thank you, Michaela!
12.04.2025 12:27 — 👍 0 🔁 0 💬 0 📌 0
Thanks!
12.04.2025 11:31 — 👍 0 🔁 0 💬 0 📌 0
Finally, we selected a representative fragment of chromatin, modelled it including histone tails, and performed all-atom MD simulations on it. The simulation showed an overall stable molecular arrangement with H4 tails mediating a stacking interaction.
11.04.2025 09:03 — 👍 14 🔁 2 💬 1 📌 0

Examining the 3D heterochromatin organization, we found a 37 nm edge-to-edge distance between connected nucleosomes and two trinucleosome arrangements: a smaller stacked population and a larger one with longer nucleosome distances, suggesting no strict long-range order for native heterochromatin.
11.04.2025 09:03 — 👍 8 🔁 0 💬 1 📌 0
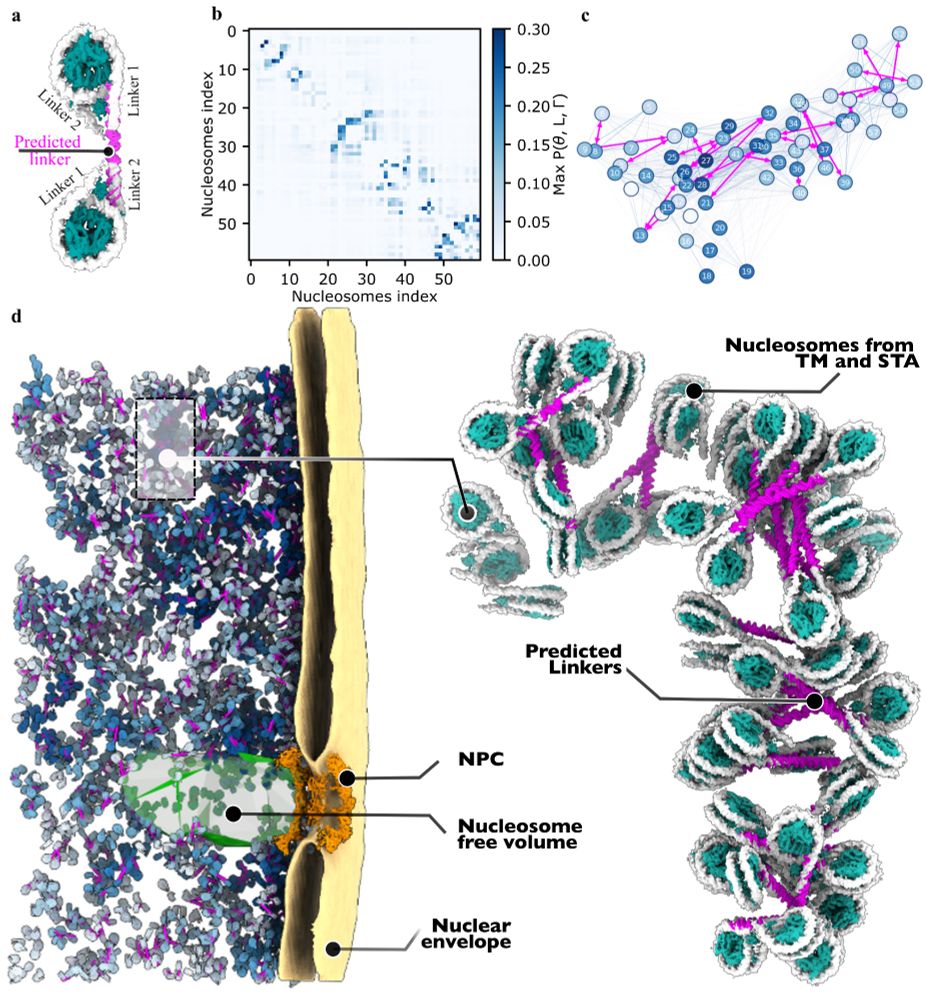
We then used a physics-based model to predict DNA linkers between the nucleosomes obtained by cryo-ET. This allowed us to visualize kbp-long chromatin fragments from inside human cells.
11.04.2025 09:03 — 👍 8 🔁 1 💬 1 📌 0

Meta-analysis on the nucleosome coordinates revealed dense chromatin patches with local concentrations exceeding 200 mg/mL and showed native chromatin behaves like an intermediate between liquid and gas with some short-range order.
11.04.2025 09:03 — 👍 3 🔁 0 💬 1 📌 0
Contextual analysis of particle data in cryo-ET #teamtomo 👨💻
Enthusiast of geometry and topology in service of understanding data 📐
PhD student at the Max Planck Institute of Biophysics (@mpibp.bsky.social) 🔬
PhD student in Eugene Kim Lab @ MPI of Biophysics
PhD student in Beckmann lab at LMU/CryoEM/Ribosome/Broad interest in structures/
Joint PhD Student at the Max-Planck-Institutes for Biophysics and -chemistry
#TeamTomo #CryoEM #Microscopy #Microfluidics #Vitrification
Postdoc @mpibp.bsky.social @wilflinglab.bsky.social studying quality control & turnover of nuclear pore complexes. Alumna @yale.edu @schliekerlab.bsky.social and @rug.nl Kampinga lab
PhD student @WilflingLab @MPIbp
archaea, methane, microbial ecophysiology, single cell activity, next generation physiology / for evidence-based decision-making / I hold strong opinions / I don’t mince words / posts reflect personal views / www.environmental-microbiology.com
Assistant Professor @UT Austin | Structural Virology | cryo-EM and cryo-ET | Postdoc with John Briggs @MRC_LMB @MPI_biochem | PhD with Elizabeth Wright @GeorgiaTech @Emory
Biochemist fascinated by visualizing biological processes with the finest spatiotemporal detail; Understand the human immune system to guide novel approaches for combating viral infections and cancer; Postdoc at Cissé Lab / MPI-IE
Membrane protein biogenesis; MPI of Biophysics
Group leader @mpi_nat, Göttingen, DE, exploring mechanisms of membrane trafficking. Cryo-EM, Cryo-ET, structural biology, cell biology.
All opinions are my own.
Biochemistry. Structural Biology in all its possible flavours. University of Göttingen. University Medical Centre Göttingen. Previously @MPI Dortmund and @MPI _Biochem.
Postdoc at the Schuller Lab - Synmikro Marburg 🇩🇪
Cryo-EM/ET of anaerobic CO₂ fixation
PostDoc in Lucas Lab at UC Berkeley. Formerly PhD student in Russo group at MRC-LMB and Master's student in Garman group at Univ. Oxford.
Cryo-ET of Rubisco phase separation
PhD @ Engel & Hondele Lab Biozentrum Basel 𝗖𝗛
#TeamTomo
Post doc in Grotjahn Lab @scrippsresearch. Mito explorer and music lover. She/her.
Scientific illustrator at https://luminous-lab.com/ and former scientist with a PhD in (bio)chemistry. Teaching design for scientists.
Tutorials: http://youtube.com/c/LuminousLab
Molecular animation at MPI for Biochemistry, Germany
In situ structural biology enthusiast | chasing molecular machines in their native habitat #cryoET Postdoc @ParkLab Scripps