While this work presents a first step towards 'decoding the repeatome' in human genome, it also presents a fast an accessible framework to assess stimulatory potential of transcripts coming from various experimental datasets.
04.10.2025 21:49 — 👍 0 🔁 0 💬 0 📌 0
Complementary to this hypothesis, it is also possible that maintaining these patterns is also beneficial for the repeat families, as we argue in the case of the highly studied family of LINE-1 repeats, where high CpG content might help prevent their deleterious effect on organism's fitness.
04.10.2025 21:49 — 👍 0 🔁 0 💬 1 📌 0
We argue that these repeats might have been co-opted to trigger innate immune system response upon translational dysregulation, such as in cancer cells. We checked experimentally that repeat transcripts that we marked as anomalous do bind to pattern recognition receptors of innate immune system.
04.10.2025 21:49 — 👍 0 🔁 0 💬 1 📌 0
It allows us to quantify how anomalous given sequence is with respect to what one would expect from a typical transcript from human genomes. The two chosen motifs, CpG content and double-stranded regions,are hallmarks of RNA viruses and our immune system recognizes them as immunostimulatory targets.
04.10.2025 21:49 — 👍 0 🔁 0 💬 1 📌 0
In our work, we instead focus on developing statistical mechanics-based framework to identify anomalous sequence (CpGf) and structure (long double-stranded segment) motifs in transcripts of these regions, which are known to be recognized by innate immune system receptors.
04.10.2025 21:49 — 👍 0 🔁 0 💬 1 📌 0
A majority of our genome does not code for proteins nor has any established regulatory function. These regions are sometimes called 'junk DNA'. Their function, or lack of, is a very active topic of research.
04.10.2025 21:49 — 👍 0 🔁 0 💬 1 📌 0
On this first day of #DNA31, reminder for the mol pro community that I put together this feed which picks up the last 3 days of posts from our community! Enjoy the meeting! 🧬
25.08.2025 08:12 — 👍 1 🔁 3 💬 0 📌 0
A great effort to chart possible future paths for the molecular programming field
17.07.2025 07:43 — 👍 1 🔁 0 💬 1 📌 0
The predictions were verified experimentally. Our results have implications for DNA/RNA molecular computing, design of DNA computing systems that interface with RNA triggers, and as we show in our presented model, it can also have implications for predicting the kinetics in CRISPR-based systems.
07.06.2025 11:05 — 👍 0 🔁 0 💬 0 📌 0
It shows that just by permuting the distribution of bases in the duplex (while keeping AT / CG base pair number constant), the kinetics of the reaction can be altered by orders of magnitude, stemming from the details of differences in stability between hybrid and canonical bases in DNA/RNA systems.
07.06.2025 11:05 — 👍 0 🔁 0 💬 1 📌 0
It is also of importance for biological systems like CRISPR-Cas9. In this collaboration, Eryk Ratajczyk supervised by Louis, Doye and Turberfield groups from Oxford, uses our new oxDNA-oxRNA hybrid coarse-grained model to study the strand displacement RNA invades DNA duplex.
07.06.2025 11:05 — 👍 0 🔁 0 💬 1 📌 0
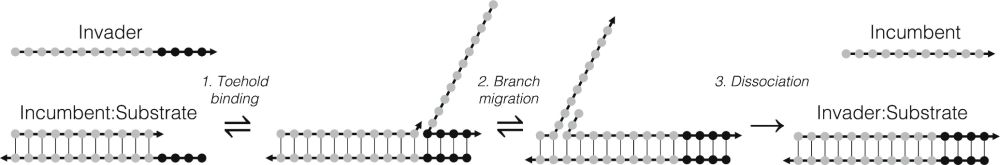
Controlling DNA–RNA strand displacement kinetics with base distribution | PNAS
DNA–RNA hybrid strand displacement underpins the function of many natural and engineered
systems. Understanding and controlling factors affecting D...
🧪🧬
Our new paper is just out in PNAS:
www.pnas.org/doi/10.1073/...
It is about nucleic acid strand displacement, which is a key reaction in molecular computation, and part of the success of the oxDNA model comes from the fact it can efficiently simulate this process.
1/4🧵
07.06.2025 11:05 — 👍 12 🔁 2 💬 1 📌 0
Great work by our student Navraj and Subhajit, building on top of the ground work of previous database by @floppleton.bsky.social and Michael Matthies
26.04.2025 22:24 — 👍 1 🔁 0 💬 0 📌 0
Nanobase
A repository for DNA/RNA nanotechnology
Just in time for #FNANO conference in Snowbird, UT, our updated and improved version of nanobase.org server is online. Just like the PDB database helped to share protein structures, we want to create a community resource for the DNA/RNA/protein nanotechnology design researchers to share their work
26.04.2025 22:23 — 👍 9 🔁 0 💬 2 📌 1
4/4: Our crystal example thus provides a falsifiability test for classical nucleation theory, showing need for also taking into account structural fluctuations.
Excellent work by Camilla Beneduce and Diogo Pinto from John Russo's group in Rome!
08.04.2025 18:00 — 👍 0 🔁 0 💬 0 📌 0
3/4 However, we find that one of the three possible crystal phases is strongly preferred, due to the crystal lattice ordering being similar to the local ordering in the liquid phase.
08.04.2025 18:00 — 👍 0 🔁 0 💬 1 📌 0
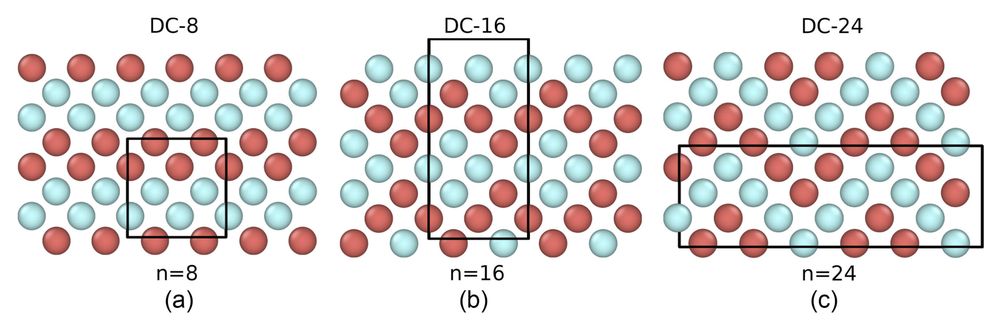
2/4: The assumptions of CNT rely on the free energy difference between bulk phase and the melt, and the interfacial free energy. In this work, we look at a two component system that forms a polymorph: it can nucleate into three different lattices with same interfacial and bulk free-energy.
08.04.2025 18:00 — 👍 0 🔁 0 💬 1 📌 0
Falsifiability Test for Classical Nucleation Theory
Classical nucleation theory (CNT) is built upon the capillarity approximation, i.e., the assumption that the nucleation properties can be inferred from the bulk properties of the melt and the crystal....
🧪In our new paper (journals.aps.org/prl/abstract...) in collaboration with Russo, Romano, Rovigatti and Sciortino groups in Rome / Venice, we look at Classical Nucleation Theory: a popular model of nucleation process, n is a key phenomena in self-assembly, self-organization and phase transitions.
08.04.2025 18:00 — 👍 6 🔁 2 💬 1 📌 0
Thank you Ulrich, it was a pleasure visiting Heidelberg and meeting you. Many thanks to the amazing group of @kgoepfrich.bsky.social for hosting me!
24.03.2025 22:31 — 👍 3 🔁 0 💬 0 📌 1
Great collaboration with @laklab-tubs.bsky.social , simulations carried out by my student Josh Evans
17.03.2025 12:42 — 👍 3 🔁 0 💬 0 📌 0
If you change something in the candano file, it has to be loaded again from scratch into oxview. The scadnano tool has recently been expanded to also allow for visualization in embedded oxview window. Having an interactive cadnano-like interface inside oxView is currently on our TBD wishlist
05.02.2025 21:15 — 👍 2 🔁 0 💬 1 📌 0
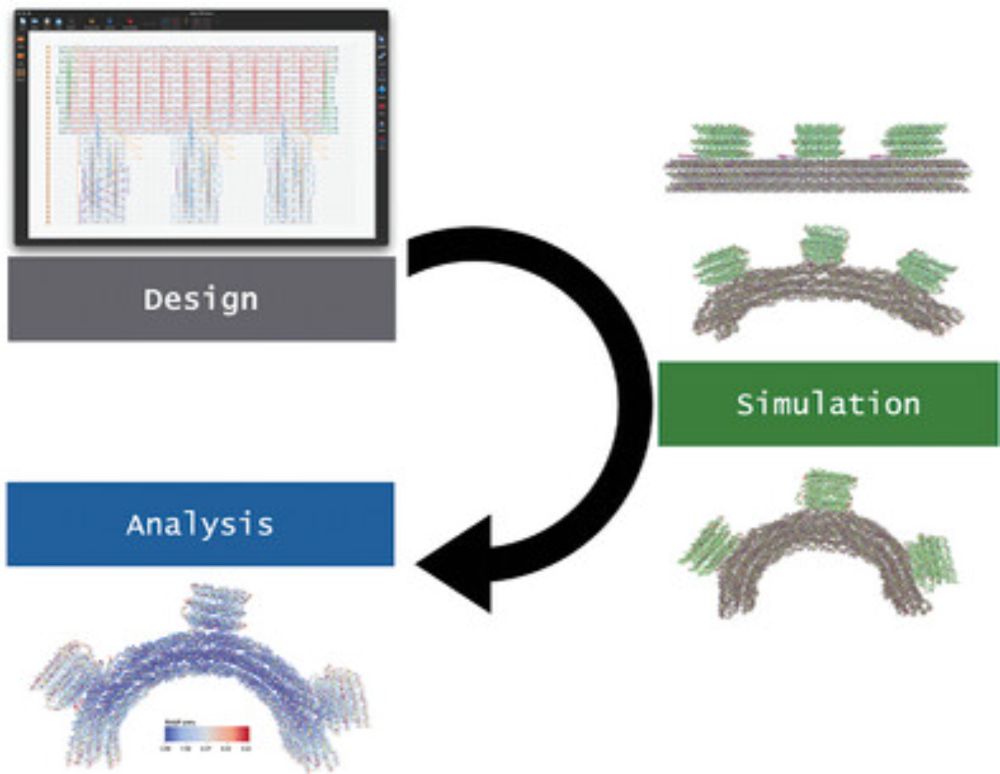
How We Simulate DNA Origami
This tutorial provides a detailed step-by-step description on how to import DNA origami designs into the oxDNA model and characterize the origami's properties (such as equilibrium shape and flexibili...
🧬 A new tutorial article from our student Sarah, walking step by step through DNA origami simulation using oxDNA model, with the help of oxview.org and oxdna.org free tools. No prior experience assumed, and also accompanied by a video: youtu.be/5-rgMekX8gE
onlinelibrary.wiley.com/doi/10.1002/...
05.02.2025 10:57 — 👍 16 🔁 2 💬 2 📌 1
Thank you!
20.01.2025 11:39 — 👍 0 🔁 0 💬 0 📌 0
Thank you!
20.01.2025 11:39 — 👍 1 🔁 0 💬 0 📌 0

Ceny Neuron 2024: Seznamte se s letošními laureáty | Nadace Neuron
Věda je plná emocí a práce vědce představuje rébus, hru, touhu po poznání, vášeň, radost i lásku. Prozkoumejte krásu těchto emocí prostřednictvím laureátů Ceny Neuron.
I am very grateful to Neuron Foundation, considered among the most prestigious awards for Czech Scientists, for awarding me Promising Scientist award in Physics yesterday at a ceremony in Prague. Many thanks goes to my family, collaborators and students: www.nadaceneuron.cz/novinky/ceny...
20.01.2025 10:03 — 👍 9 🔁 0 💬 2 📌 0
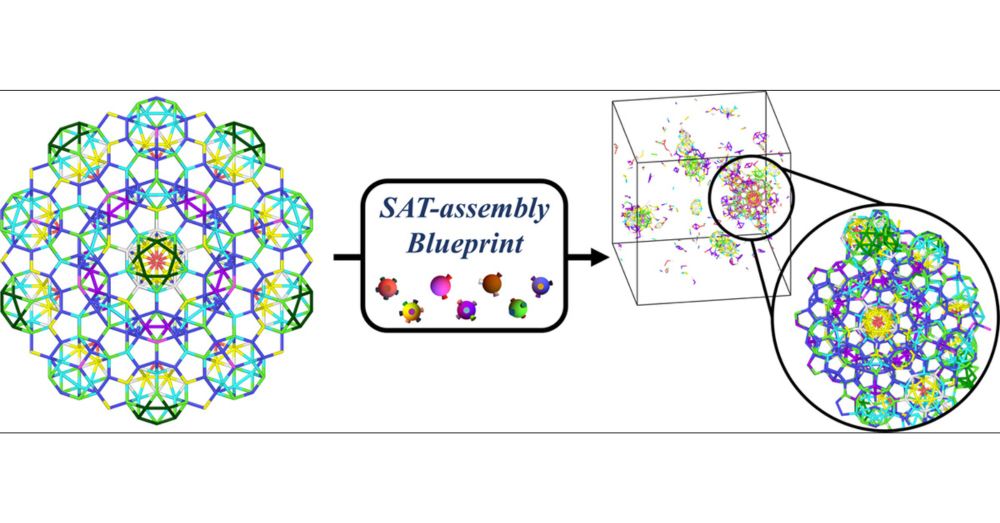
Automating Blueprints for the Assembly of Colloidal Quasicrystal Clusters
One of the frontiers of nanotechnology is advancing beyond the periodic self-assembly of materials. Icosahedral quasicrystals, aperiodic in all directions, represent one of the most challenging target...
🧪🧬 New article with J Russo, D. Pinto and F. Sciortino out is @acspublications.bsky.social ACS Nano: pubs.acs.org/doi/10.1021/...
We use our #SAT Assembly method to design particles that assemble into 3D quasicrystal clusters, paving a way to their experimental realization with DNA nanostructures
22.12.2024 08:48 — 👍 13 🔁 5 💬 0 📌 0
YouTube video by Sapienza soft matter group
SATa CLAUSE: When magic snow falls, a enchanted meadow blossoms
In our Christmas tradition with John Russo's lab at Sapienza university of Rome, here is a Christmas-themed video highlighting a current research project based on our #SAT-assembly method, this time chiral crystals self-assembly (made by C. Beneduce): www.youtube.com/watch?v=0A7x...
13.12.2024 01:13 — 👍 3 🔁 0 💬 0 📌 0
Glad you find it useful!
03.12.2024 16:26 — 👍 1 🔁 0 💬 0 📌 0
Postdoctoral Fellow & DNA/RNA Origami facility lead at Universität Heidelberg | Göpfrich group | DNA/RNA Origami | PhD from TUM | Dietz Lab | DNA Origami
Chemist, 🧲 spectroscopist, mother, Associate Teaching Prof. of Chemistry at University of Denver. Loves reading romance and nonfiction 💖📚, currently writing a narrative nonfiction book on spectroscopy🚨Opinions and thoughts and posts are my own!
Tinnefeld lab @lmumuenchen.bsky.social 🧬
Biophysicist working on lipid membranes & drug delivery. Currently postdoc in the Weitz Lab @Harvard - PhD @mpi_mr_hd with @KGoepfrich.
Inventing the future of biology with chemistry and biophysics. EMBO Long-Term Fellow at the Szostak Lab, HHMI & University of Chicago. Former Winton, Schiff, and Benefactors’ Scholar at the University of Cambridge’s Keyser Lab. 🧬🧪🕳️🔬
Physicist - Doctoral Candidate @jungmannlab.bsky.social
Exploring the cellular world at the nanometer scale with DNA-PAINT.
@mpibiochem.bsky.social
@lmumuenchen.bsky.social
Minister Spraw Zagranicznych 🇵🇱🇪🇺
Professor Université de Montréal, Physics of Living Systems, Machine Learning, etc…
https://www.francoisresearch.org/
Theoretical physicist interested in the physics of living systems and statistical physics. Professor at LMU Munich. Passionate about emergent phenomena and interdisciplinary research.
https://www.theorie.physik.uni-muenchen.de/lsfrey/
Computational structural biologist @Fox Chase Cancer Center. Gay, out & proud since 1986. I support transgender & nonbinary people. Living with CLL/SLL. Video about my LGBTQ+ life in science: https://tinyurl.com/45srzjdw. Views my own, not my employer's.
Prof Biomolecular Simulations Uni Mainz. Disordered proteins, nucleic acids and their dynamics, complexes and phase-separated condensates. Integrating simulations and experiments.
Physicist and engineer, into multiple scale models of proteins/RNA/membranes, polymers, scientific computing, more --> http://tinyurl.com/goCompModeling
Computational biophysics, cell motility, collective motion, soft matter, horses, cats. Associate Prof at Johns Hopkins Physics+Biophysics departments.
Laboratory of Physical and Biological Chemistry of Living Matter (UMR 8228)
ENS, CNRS, Sorbonne University, Paris
Assistant Professor in Chemistry at Case Western Reserve University.
Group of passionate RNA biologists in the Rhineland area in Germany (Cologne/Bonn/Düsseldorf/Aachen). Sponsored by the RNA Society and Lexogen. Run by @inahuppertz.bsky.social and @katleppek.bsky.social
An international high impact journal exploring the science of biomaterials and their translation towards clinical use.
Published by @rsc.org 🌐 Website: rsc.li/biomatersci