 02.12.2025 14:56 — 👍 0 🔁 0 💬 0 📌 0
02.12.2025 14:56 — 👍 0 🔁 0 💬 0 📌 0
@inversion.bsky.social
Genomics nerd.
@inversion.bsky.social
Genomics nerd.

My new tool Paraviewer is now available for use at github.com/PacificBiosc...! If you use Paraphase, try this new next-step tool - it automates and greatly simplifies variant visualization from Paraphase variant calling. If you're at #ASHG2025, visit me today at poster 4109W. #pacbio
15.10.2025 14:58 — 👍 7 🔁 5 💬 1 📌 0
#PacBio announces major upcoming advances for #Revio and #Vega designed to lower genome costs, expand multiomic capabilities, and support regulated research.
The SPRQ-Nx chemistry, now in beta access, delivers complete genomes for under $300 at scale.
View the press release: bit.ly/4o06eyH

My complex variant visualization tool SVTopo is now officially published in BMC Genomics! link.springer.com/article/10.1.... This tool allows HiFi users to view complex germline structural variation in intuitive and informative plots.
09.10.2025 19:00 — 👍 6 🔁 7 💬 2 📌 0It absolutely warms my heart when I click on a pre-print that includes a bunch of top methods folks from industry and I see (1) open-source software (with a good license; MIT in this case) that is (2) written in Rust. Bravo @pacbio.bsky.social!
Moar Rust plz!
github.com/PacificBiosc...

Arthur continues, showcasing how long-read sequencing discovered a novel fusion in an ovarian cancer patient—one that short-read methods miscalled as simple overexpression of the 3' fusion partner. #AACR25 #PacBio
28.04.2025 18:20 — 👍 3 🔁 3 💬 0 📌 0
Interesting work from Khi Pin Chua, PhD on precise characterization of complex repeat regions in cancer genomes at #AACR25. Catch him at poster 2 in section 46 (#6288) until 5 PM! #PacBio
29.04.2025 19:29 — 👍 4 🔁 3 💬 0 📌 0
Advanced genomic analysis of 4 generation family offers new knowledge about genetic mutations & their transmission, including inherited variants & those that arise anew
@nature.com @uwgenome.bsky.social @eichlerlab.bsky.social @uwmedicine.bsky.social @utah.edu @pacbio.bsky.social

Long-read sequencing of large pedigrees is an ideal way to map all classes of denovo mutations! A collaboration between University of Utah, University of Washington, and PacBio. Glad to be a part of this project 👏
www.nature.com/articles/s41...
Really neat paper out, using measurements of 3D genome folding (with our Proximo Hi-C tech, wink wink) to pinpoint the CA2 gene as a potential target to reverse drug resistance in #breastcancer.
22.04.2025 15:42 — 👍 1 🔁 1 💬 0 📌 0
I just released a new preprint! The manuscript describes SVTopo, a software tool that enhances visualization of complex SVs using HiFi data: www.biorxiv.org/content/10.1.... Here’s a summary of the results:
22.04.2025 14:21 — 👍 13 🔁 8 💬 1 📌 1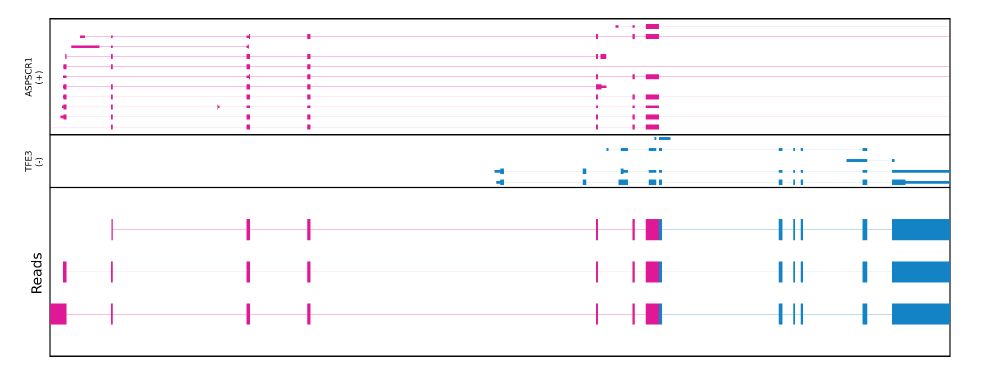
Fusion transcript plot
Checkout out a new release of pbfusion (0.5.0) for accurate detection and visualization of fusion transcripts from @pacbio.bsky.social HiFi data.
github.com/PacificBiosc...

“StarPhase: Comprehensive Phase-Aware Pharmacogenomic Diplotyper for Long-Read Sequencing Data” is now on biorxiv! In this work, we explore the use of long-read sequencing (#PacBio #HiFi) for #pharmacogenomics #PGx. 1/N
Pre-print: doi.org/10.1101/2024...
Repo: github.com/PacificBiosc...
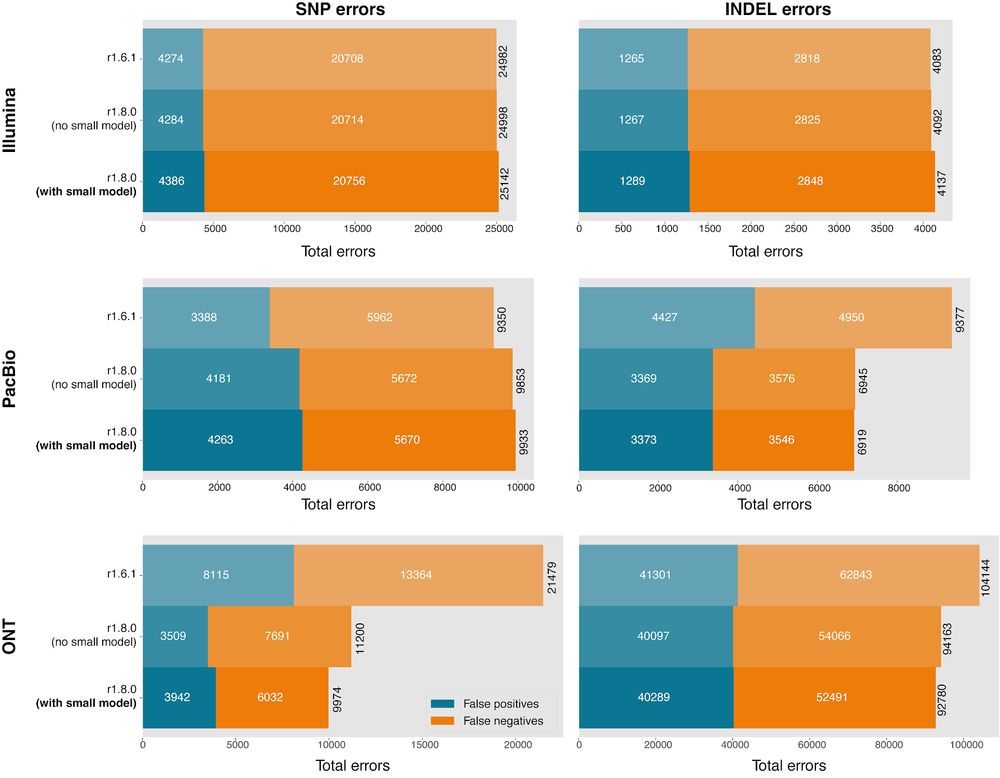
Plots of SNP and Indel error numbers for DeepVariant models. Shows a Indel error reduction of 26% for PacBio and a ~50% SNP error reduction for ONT.
Added SPRQ to PacBio training, reducing Indel error on SPRQ by 26%. Added Platinum Pedigree training data for PacBio model, reducing errors by 34% on more extensive Platinum truth. New model and case study for Kinnex/Mas-Seq/Iso-Seq. Additional speed options for GPU pipelines 2/3
05.12.2024 17:57 — 👍 8 🔁 4 💬 3 📌 0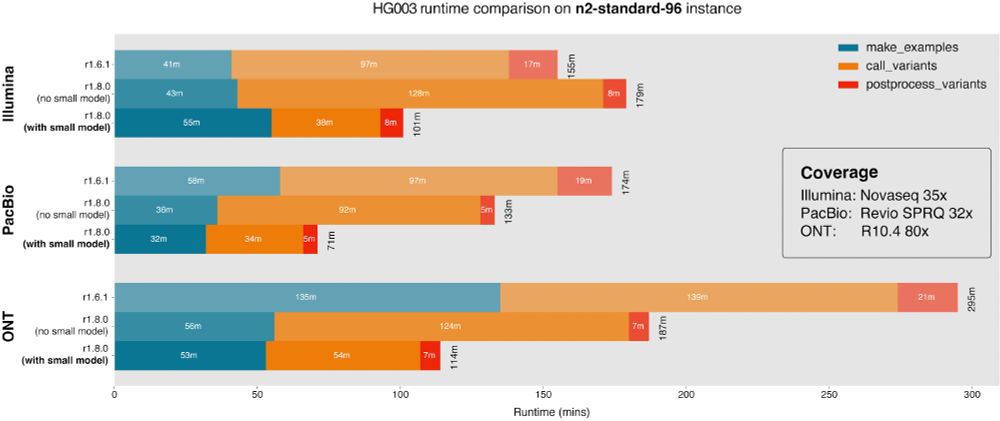
Runtime figure for new version of DeepVariant with and without small model. Showing reduction in runtime of 155 minutes to 101 minutes with Illumina, 174 minutes to 71 minutes with PacBio, and 295 minutes to 114 minutes with Oxford Nanopore.
Release of DeepVariant 1.8. Large speed improvement (~67% faster) via small model for easy sites. New Pangenome-aware option. Reduces error by ~30% for vg-mapped WGS, ~10% for BWA WGS, ~5% BWA exome. New config for custom model users, see release notes.
(github.com/google/deepv...)

Our special issue @genomeresearch.bsky.social is out on long-read sequencing (@pacbio.bsky.social & ONT): genome.cshlp.org/content/34/1...
Super excited about all the new work and special thanks to all the authors! Big thanks to Hillary @ahoischen.bsky.social @anaconesa.bsky.social
Howdy, please add me. All things genomics 😊
19.11.2024 22:39 — 👍 0 🔁 0 💬 0 📌 0
How to call SNV in RNA seq from Nanopore / Pacbio ? Easy: Clair-3RNA biorxiv.org/content/10.1... ~95% / ~96% F1-score for ONT / PacBio + we detect RNA editing! Great collaboration with Ruibang Luo, his group + Miten Jain ( need to invite them over here).
Special thanks to Justin Zook from GIAB!
The new chemistry requires less sample input, and Kinnex certainly reduces the isoSeq costs.
19.11.2024 01:56 — 👍 1 🔁 0 💬 0 📌 0
Confirmed utility of long reads in detecting fusion rearrangements in both HiFi WGS and IsoSeq.
www.medrxiv.org/content/10.1...