A first in human genomics: a 4-generation pedigree reference, now in Nature.
Built with #PacBio HiFi, it maps de novo mutation rates, reveals paternal bias, and captures high mutation rates in tandem repeats—even in Y and repeat-rich regions.
Paper here: go.nature.com/4lGMPlP
#TheresHiFiForThat
24.04.2025 15:34 — 👍 10 🔁 6 💬 1 📌 0
Oh Shit, Git!?!
For starters, the software carpentry lesson (swcarpentry.github.io/git-novice/) is pretty good. When things go wrong, or when you want to dig deeper, I like ohshitgit.com, and more generally, blog posts by Julia Evans related to git: jvns.ca#git
14.04.2025 17:26 — 👍 8 🔁 1 💬 1 📌 1

7500 research staff at the University of California filed a petition to form a union with UAW today at the Public Employment Relations Board in Oakland. While the work is really just beginning, we have had thousands of conversations to get here. We're going to win ✊
12.04.2025 00:22 — 👍 5 🔁 3 💬 0 📌 0

ESHG Conference 2025
ESHG 2022 COVID-19 Information
Discover the ESHG 2025 - Hybrid Conference programme.
We are proud to announce the confirmed 2025 speakers.
Stay updated on matters related to the ESHG soc...
Also worth noting that a substantial new sawfish CNV integration feature will be coming as a preview release on GitHub later this month, which I’ll also be detailing as a poster presentation at ESHG. Looking forward to sharing more about this soon.
10.04.2025 15:41 — 👍 2 🔁 1 💬 1 📌 0

Sawfish: Improving long-read structural variant discovery and genotyping with local haplotype modeling
AbstractMotivation. Structural variants (SVs) play an important role in evolutionary and functional genomics but are challenging to characterize. High-accu
Great to see that sawfish, our new HiFi SV caller, is accepted for publication in Bioinformatics! Sawfish emphasizes local haplotype modeling to improve SV representation and genotyping in both single and joint-sample analysis. Advance-access article now available: (1/n)
doi.org/10.1093/bioi...
10.04.2025 15:41 — 👍 44 🔁 24 💬 2 📌 1
Get in Dorks, we are going protesting.
STAND UP FOR SCIENCE. MARCH 7th 12-4pm. DC AND YOUR STATE CAPITALS.
More information to come.
09.02.2025 14:23 — 👍 856 🔁 433 💬 42 📌 38
Long-read sequencing resolves the clinically relevant CYP21A2 locus, supporting a new clinical test for Congenital Adrenal Hyperplasia https://www.medrxiv.org/content/10.1101/2025.02.07.25321404v1
11.02.2025 04:40 — 👍 3 🔁 2 💬 0 📌 0

GitHub - PacificBiosciences/pb-StarPhase: A phase-aware pharmacogenomic diplotyper for PacBio datasets
A phase-aware pharmacogenomic diplotyper for PacBio datasets - PacificBiosciences/pb-StarPhase
“StarPhase: Comprehensive Phase-Aware Pharmacogenomic Diplotyper for Long-Read Sequencing Data” is now on biorxiv! In this work, we explore the use of long-read sequencing (#PacBio #HiFi) for #pharmacogenomics #PGx. 1/N
Pre-print: doi.org/10.1101/2024...
Repo: github.com/PacificBiosc...
11.12.2024 14:30 — 👍 19 🔁 16 💬 2 📌 3

ICYMI my poster at #AMPath24, I'm sharing it here. Folks interested in long reads to resolve complex loci like repeat expansions relevant to neuro disease and carrier screening, check it out! @pacbio.bsky.social collab with @egor-dolzhenko.bsky.social @guilhermesena1.bsky.social and many others
23.11.2024 14:59 — 👍 15 🔁 8 💬 3 📌 0
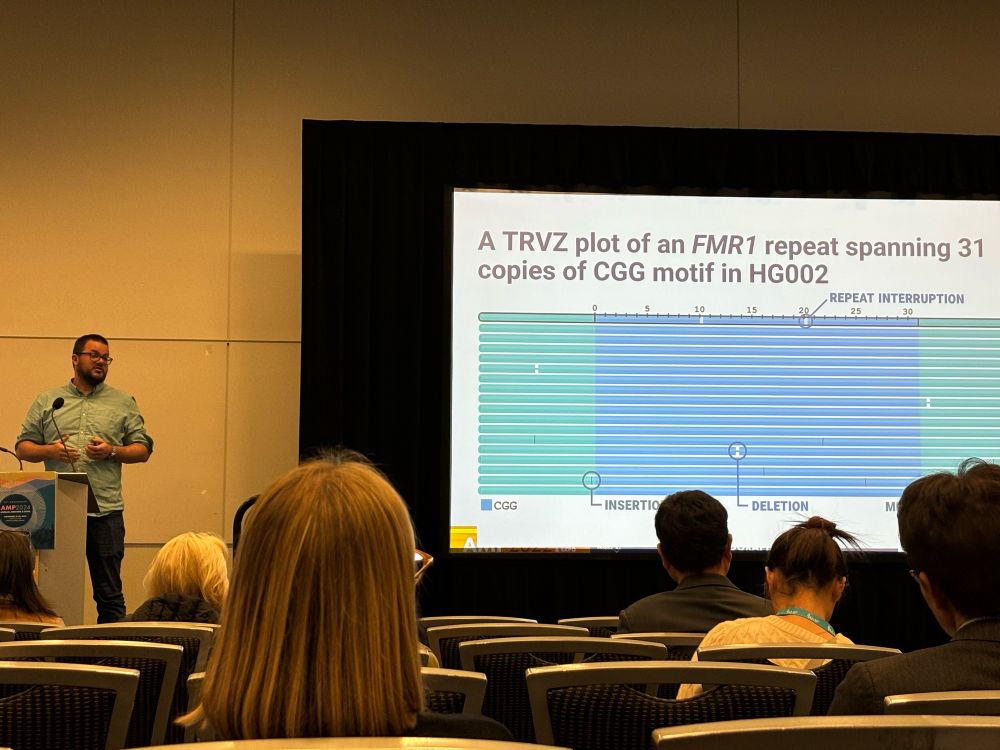
@sedlazeck.bsky.social AMPath24 talk about long reads highlighting TRGT tool by @egor-dolzhenko.bsky.social and @guilhermesena1.bsky.social and others for HiFi sequencing by @pacbio.bsky.social
23.11.2024 19:23 — 👍 17 🔁 6 💬 0 📌 0

Careers - PacBio
PacBio is always looking to add the best and brightest minds to our world-class company. Our highly interdisciplinary team is best suited for individuals who are creative, forward thinking, and who ap...
If you are a #C++ developer, either have a #bioinformatics or #CUDA background, and take pride in your engineering and/or algorithmic skills, I'm looking for a Senior Staff Engineer at @pacbio.bsky.social. You can work remotely. Feel free to ping me or go to pacb.com/careers
Please spread the word
15.11.2024 15:08 — 👍 14 🔁 7 💬 0 📌 0

We can see there's already excitement for Vega on Bluesky. ✨ What do you think about the first #PacBio HiFi benchtop system?
13.11.2024 17:46 — 👍 7 🔁 4 💬 0 📌 0
I think I'll try hiring @pacbio.bsky.social a #bioinformatics engineer with modern C++ skills. Interested? You can be remote in the US, UK, Germany, or Switzerland. Goals: implement crazy algorithms for on-instrument sequence data processing and improve existing solutions. Contact me.Spread the word
12.11.2024 23:05 — 👍 9 🔁 4 💬 0 📌 0
Waking up in my own normal body and freaking out like the guy who woke up as a bug
09.02.2024 14:58 — 👍 1658 🔁 295 💬 25 📌 16
My poster on long read pharmacogenomics with #PacBio HiFi is up for today at #ASHG24. Ping me on here if you want to meet before the poster session!
08.11.2024 16:36 — 👍 3 🔁 2 💬 0 📌 2

Interested in long-read pharmacogenomics? Then I have some exciting things to show you at #ASHG24... Looking forward to next week in Denver!
#PacBio #PGx
28.10.2024 20:08 — 👍 8 🔁 2 💬 1 📌 0
Giulia Del Gobbo #ACMGMtg24 on lessons learned with Care4Rare using #PacBio #HiFi: need platform matched controls, discovering overlooked variants, phasing aid in interpretation, need specialized STR tools, and higher resolution of structural variants.
15.03.2024 17:45 — 👍 1 🔁 1 💬 1 📌 0
Emily Farrow #ACMGMtg24 on "Unveiling the power of #HiFi genome sequencing: one test to rule them all?" Clinical validation of #PacBio Revio; overall high validation accuracy; 26/50 cases with solves; average 25 day TAT; combination evaluation of SNV, CNV, SV, and methylation.
15.03.2024 18:03 — 👍 2 🔁 1 💬 1 📌 0

Release MethBat v0.9.0 · PacificBiosciences/MethBat
Changes
Adds a new segmentation mode based on Circular Binary Segmentation. This mode generates a BED file of methylated, unmethylated, and ASM regions. The mode is executed with methbat segment ....
We just added a methylation segmentation algorithm for PacBio HiFi data to our MethBat tooling (beta). If you are interested in trying it out, we would greatly appreciate any feedback on the performance and possible extensions!
Release here: github.com/PacificBiosc...
09.02.2024 18:39 — 👍 2 🔁 2 💬 0 📌 0
Excited to announce that our accompanying paper on HiPhase has been published today, I'll highlight some of the additions in this thread! #PacBio #HiFi
“HiPhase: Jointly phasing small, structural, and tandem repeat variants from HiFi sequencing”: doi.org/10.1093/bioi...
26.01.2024 15:07 — 👍 4 🔁 3 💬 1 📌 0
Listening to _Four Thousand Weeks_ at 2x speed to save time.
13.01.2024 07:53 — 👍 2 🔁 0 💬 0 📌 0
With our outstanding partners at @GeneDx , @PacBio and @GoogleHealth, we will take on the biggest genomic diagnostic challenge of all, diagnosing rare diseases in individuals who are not only extensively tested already, but are from genetic ancestries ignored by medical genomics
09.11.2023 02:19 — 👍 3 🔁 1 💬 1 📌 0
Release HiPhase v1.0.0 · PacificBiosciences/HiPhase
Changes
Added support for tandem repeat calls from TRGT; minimum supported version - v0.5.0
VCF index files (.tbi) are now automatically generated by HiPhase
The reference FASTA file is now a requ...
I'm pleased to announce the release of HiPhase v1.0.0! In addition to phasing small and structural variants from #PacBio #HiFi datasets, HiPhase now also includes support for phasing short tandem repeats (#STR) at the same time!
Full release notes can be found here: github.com/PacificBiosc...
30.10.2023 21:45 — 👍 8 🔁 7 💬 1 📌 1

GitHub - google/deepsomatic
Contribute to google/deepsomatic development by creating an account on GitHub.
Initial release of DeepSomatic, which identifies subclonal variants when given tumor and normal BAM files. Pre-trained models and case studies available for Illumina and PacBio. Development led by Kishwar Shafin which built off a framework by Pi-Chuan Chang.
(github.com/google/deeps...)
24.10.2023 16:53 — 👍 36 🔁 17 💬 2 📌 1

Release DeepVariant 1.6.0 · google/deepvariant
Improved support for haploid regions, chrX and chY. Users can specify haploid regions with a flag. Updated case studies show usage and metrics.
Added pangenome workflow (FASTQ-to-VCF mapping with V...
Release of DeepVariant v1.6.
Support for haploid regions, chrX/Y.
Workflow for Pangenome FASTQ-to-VCF.
Major DeepTrio improvements for de novo variants.
Models for CompleteGenomics T7, G400
Add NovaSeqX to training data
Release by Kishwar Shafin
github.com/google/deepv...
26.10.2023 16:32 — 👍 7 🔁 4 💬 1 📌 0
Decoding how the gut thinks 🦠🪱🧠💪
Neuroscientist with interests in
#EnergyMetabolism #EntericNeurons #Fats
@crick.ac.uk @institutducerveau.bsky.social
Entertaining & educational conversations about science, tech, + more. Hosted by Ira Flatow and Flora Lichtman. From WNYCStudios.
Algae enthusiast, Associate Professor @UniofExeter, joint appointment @thembauk
Algal ecophysiology | signalling | microbiome | molecular microbiology | diatoms!
Clinical Psych PhD candidate at Emory
Founder and CEO of @standupforscience.bsky.social
Standupforscience.net
"Colette is wearing brass knuckles...w/ spikes tipped in poison"
“Delusional broad”
"Lady Switchblade"
“Ditsy socialist liberal”
Mobilizing the fight for science and democracy, because Science is for everyone 🧪🌎
The hub for science activism!
Learn more ⬇️
http://linktr.ee/standupforscience
Poet. Opinions are my own
Researching somatic evolution at the University of Cambridge and Wellcome Sanger Institute. Also a scientific illustrator.
Community Notes for Science. Tag @ScholarAI anywhere on Bluesky Social to get evidence-based answers automatically. Follow for science updates.
Head of Research Department @uclqsneuromuscular.bsky.social | Professor of Clinical Neurology and Mitochondrial Medicine @uclqsion.bsky.social | Lead @mrcmitocluster.bsky.social | Co-lead @londonmito.bsky.social
The Genomic Health Record unifying and standardizing genetic care, complete with #pedigree drawing, risk assessment, #HPO phenotyping, and more.
Associate Professor, University of Arizona
Algorithms, Machine Learning, Data Resources
Computational Genomics, Drug Discovery, Animal Tracking, etc.
wheelerlab.org
Focused on innate immune mechanisms governing pathogenesis of disease and on NK-cell based therapies for disease. Opinions expressed are Dr. Waggoner's alone and not on behalf of lab staff or employer.
Group leader of transposon group in Oxford at IDRM. #new_pi https://sites.google.com/view/berrenslab
Palaeoanthropologist at the Natural History Museum London.
Posts are my own, and when I repost or like, it does not signify I necessarily agree with all content https://www.nhm.ac.uk/our-science/departments-and-staff/staff-directory/chris-stringer.html
study small RNAs/APA/splicing/m6A/Notch. neural development+behavior; genomic conflict+evolution.
http://www.mskcc.org/lai
also, indie rock and glassblowing.
Building a home base for bioinformatics in the era of biology.
Visit our website at bioinformaticsbase.com or bioinformaticsbase.substack.com to read our articles.
MSCA Doctoral Candidate | @progret2024.bsky.social | debaerelab.com | UGent | CMGG 🇧🇪
Studying Retinal Dystrophies and 3D Genome 👁️🌀
@eshgyoung.bsky.social
Alumni from Carla Oliveira Lab (i3S,🇵🇹), Caldas Lab (CRUK CI, 🇬🇧) & Ana-Teresa Maia Lab (UAlg, 🇵🇹)


















