
Congrats to the leading authors Baptiste Gaborieau,
Hugo Vaysset, and @ftesson.bsky.social and to all the great team.
www.biorxiv.org/content/10.1...
@mdmlab.bsky.social
Molecular Diversity of Microbes Lab at Institut Pasteur Focusing on bacterial immunity 🦠🧬🧫 // We ♥️ sharing our science with everyone // PI Aude Bernheim

Congrats to the leading authors Baptiste Gaborieau,
Hugo Vaysset, and @ftesson.bsky.social and to all the great team.
www.biorxiv.org/content/10.1...
So, can we predict phage-bacteria from their genomic data ? Yes, to a certain extent !
There is much more work to be done to improve these predictions, but our study demonstrates feasibility. We anticipate similar approaches could be applied to many bacterial species.
We are amazed by the diversity of phage-bacteria interactions beyond laboratory models 🤩 .
We hope that making available to the community 2 new collections & the accompanying interaction dataset will provide a starting point for mechanistic exploration of these interactions.
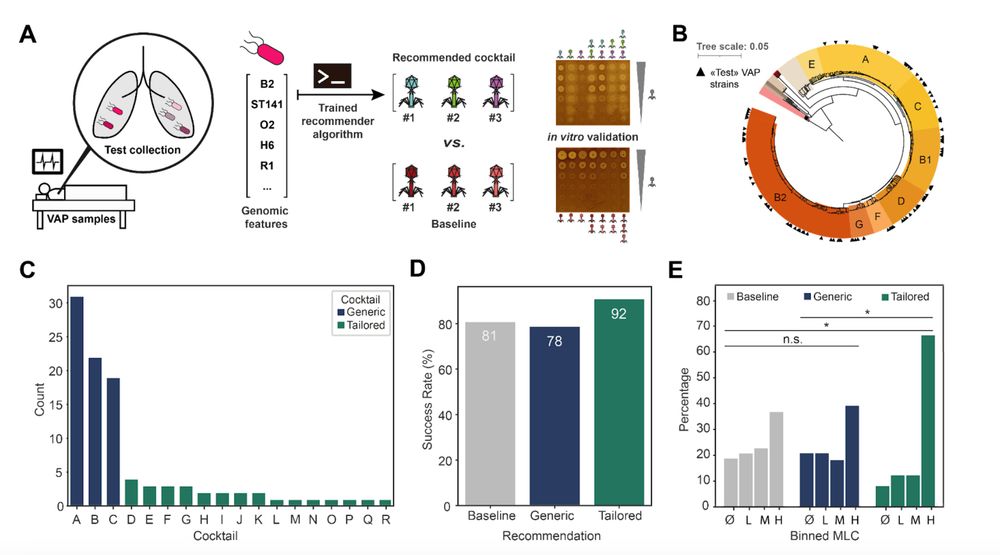
Finally, we show the application of such predictions by establishing a pipeline to recommend tailored phage cocktails to target pathogenic strains from their genomes only🧬💻.
We show higher efficiency of tailored cocktails on a collection of 100 pathogenic E. coli isolates.
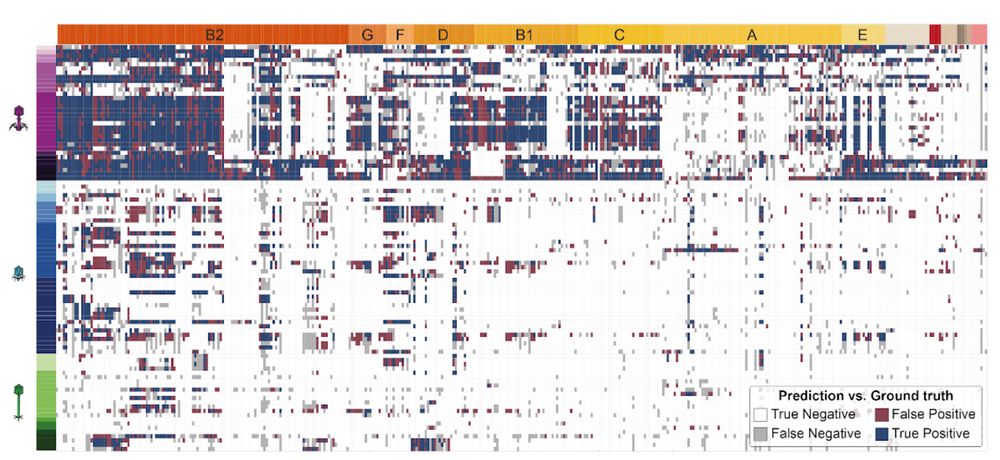
We then trained predictive algorithms to figure out which interactions we can accurately predict. We got an overall AUROC of 86%. We provide a roadmap for the future as it will allow us to focus on the 3,379 false negatives and 2,922 false positives that are not well predicted.
23.11.2023 08:00 — 👍 4 🔁 0 💬 0 📌 0On the bacterial side, we show that most interactions in our dataset can be explained by adsorption factors as opposed to antiphage systems which play a marginal role (more about antiphage systems in the manuscript).
23.11.2023 07:59 — 👍 2 🔁 0 💬 0 📌 0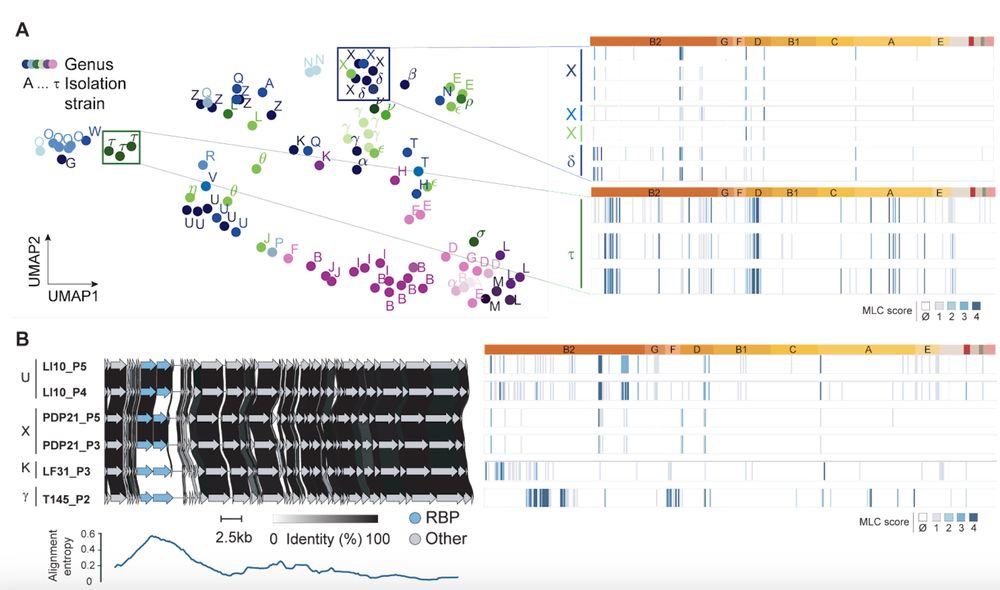
On the phage side, the strain the phage was isolated on accounts for 53% of the variance (!). We show this is linked to the phage ability to adsorb which is driven by its Receptor Binding Proteins. Side note: Only ⅓ of out phages were able to infect Coli K-12
23.11.2023 07:59 — 👍 2 🔁 0 💬 0 📌 0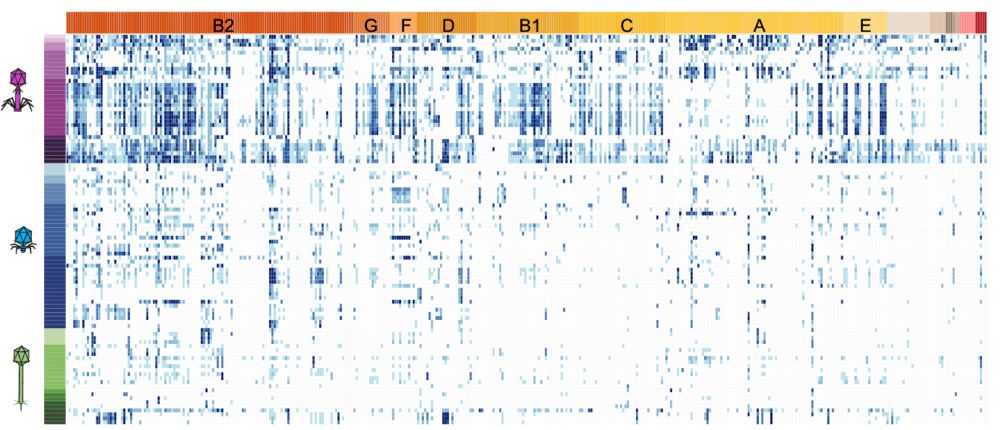
Then came the team effort 🤩. We infected everyone against everyone generating a matrix >38000 interactions. This was done in triplicates with 3 concentrations. The matrix of interactions reveals complex (and beautiful !) patterns. How much of these do we understand ?
23.11.2023 07:58 — 👍 1 🔁 0 💬 0 📌 0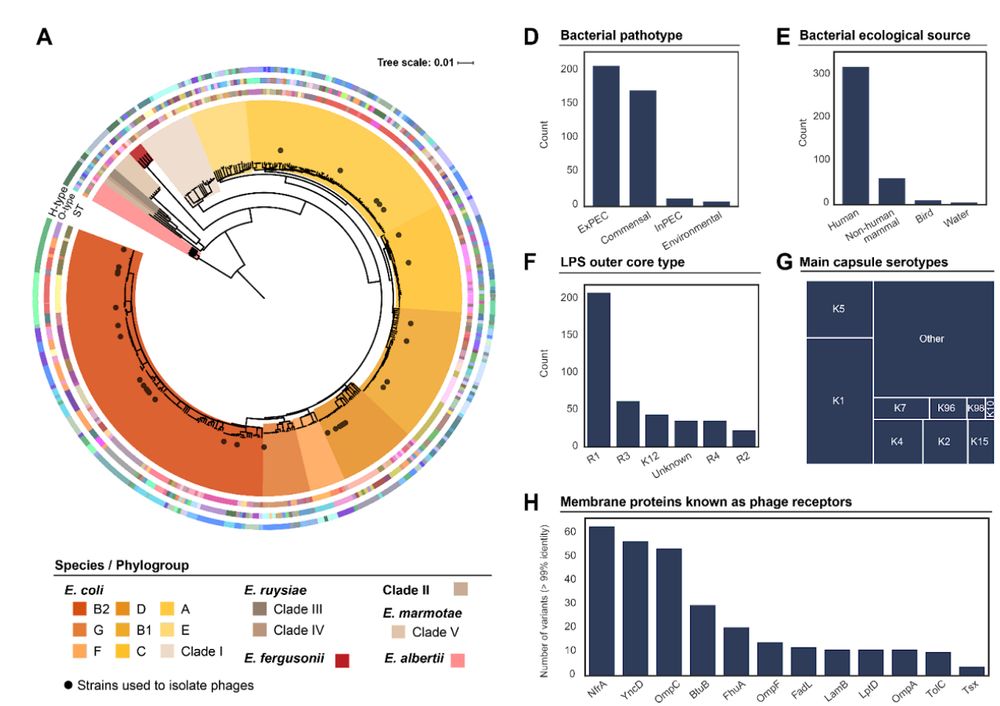
We needed a dataset, diverse and at scale. We chose the Escherichia genus. We established 2 collections of 403 natural, diverse, Escherichia strains and 96 bacteriophages. We looked in their genomes 💻🧬 for traits related to infection (receptors, capsules, defense systems…).
23.11.2023 07:57 — 👍 2 🔁 0 💬 0 📌 0Recent advances in genomics enable the identification of traits linked to phage-host specificity, suggesting the potential use of these traits for predicting such interactions. So how well can we predict a range of phage-bacteria interactions exclusively from their genomic data?
23.11.2023 07:55 — 👍 0 🔁 0 💬 0 📌 0Predicting which phages infect specific strains would allow to better fight bacterial infections and understand microbial ecology . Many studies focused on model phage-bact couples, how concepts learns from these apply to the *super* diverse natural context remains uncertain.🤔
23.11.2023 07:54 — 👍 1 🔁 0 💬 1 📌 0💻🧫 New preprint: How accurately can we predict diverse phage bacteria-interactions from their genomes only ? We created a matrix of >38k phage-bacteria interactions to find out (=> AUROC 86%) & used our predictions to recommend tailored phage cocktails.
www.biorxiv.org/content/10.1...