10 days until the deadline!
05.12.2025 13:32 — 👍 3 🔁 1 💬 0 📌 0

illustration of petri dishes growing different microbes
We're excited to help launch the inaugural Gordon research conference on #microbiome editing, coming up in January in Pomona, CA! Learn more and register here: https://ow.ly/a6Am50X9XkE
27.10.2025 15:05 — 👍 19 🔁 9 💬 2 📌 2

Engineered probiotic restores GLP-1 signaling to ameliorate fiber-deficiency exacerbated colitis
Engineered probiotic delivers bacterial peptide that enhances GLP-1 and restores barrier integrity in diet-modulated colitis.
Highlighting another recent paper from the lab in Science Advances on engineering a gut probiotic to modulate GLP-1 levels and ameliorating colitis. Led by Leonie Brockmann & @carlottaronda.bsky.social . @columbiasysbio.bsky.social @columbiauniversity.bsky.social 🧬🦠💊
www.science.org/doi/full/10....
21.11.2025 01:32 — 👍 8 🔁 3 💬 0 📌 0
We are excited for many apps of Foli-seq in academia & industry. Please reach out to collaborate! Lots of exciting projects in this space for students & postdocs. Led by Yiming @yiminghuang.bsky.social and Yiwei @yiweisun.bsky.social & thanks to all our collaborators!
@columbiasysbio.bsky.social
17.11.2025 20:55 — 👍 2 🔁 1 💬 0 📌 0
In 💩 samples from patients with Inflammatory Bowel Disease (IBD) from the @crohnscolitisfdn.bsky.social, we can distinguish different patient cohorts with increase clinical severity of disease and stratifying biomarkers that associated with immune pathway involvement.
17.11.2025 20:55 — 👍 0 🔁 0 💬 1 📌 0
Each day, humans shed billons of host cells from the gut lining out of the body (our "exfoliome") via our poop. Foli-seq unlocks this non-invasive exfoliome analysis to assess gut and immune functions, and links with the gut microbiome, all w/o colonoscopy or tissue biopsy. 💩🧬
17.11.2025 20:55 — 👍 0 🔁 0 💬 1 📌 0

Fecal exfoliome sequencing captures immune dynamics of the healthy and inflamed gut - Nature Biotechnology
Profiling host RNA shed in feces reveals disease states in the gut.
Introducing Foli-seq in @natbiotech.nature.com today. Foli-seq can be used to measure gut-derived RNA from fecal matter (signals of immune, secretory & epithelial cells). We used this to map gut inflammation, drug response, and host-microbe interactions. 💩🧬💊🦠
www.nature.com/articles/s41...
17.11.2025 20:55 — 👍 19 🔁 12 💬 1 📌 0

📢 Our Dept. of Systems Biology at Columbia University has an open tenure-track Assistant Professor position in the broad area of quantitative biology. Come join our awesome department in NYC! Please circulate.
apply.interfolio.com/177622
Suggested deadline: 12/15/2025.
@columbiasysbio.bsky.social
15.11.2025 04:02 — 👍 31 🔁 37 💬 0 📌 1
2026 Microbiome Editing Conference GRC
The 2026 Gordon Research Conference on Microbiome Editing will be held in Pomona, California. Apply today to reserve your spot.
We are hiring PhD & Postdocs in this space so please reach out if interested. Also check out the Gordon Research Conference on Microbiome Editing in Jan 11-16, 2026.
www.grc.org/microbiome-e...
13.11.2025 20:53 — 👍 6 🔁 2 💬 1 📌 0
Led by the stellar Diego Gelsinger @drgel.bsky.social working with many contributors through a wonderful long-term collab w/ Sam Sternberg & Ivo Ivanov labs! 🤝 @columbiauniversity.bsky.social
@columbiasysbio.bsky.social
13.11.2025 20:53 — 👍 4 🔁 0 💬 3 📌 0
Congrats Bryan!!
05.05.2025 21:05 — 👍 1 🔁 0 💬 0 📌 0
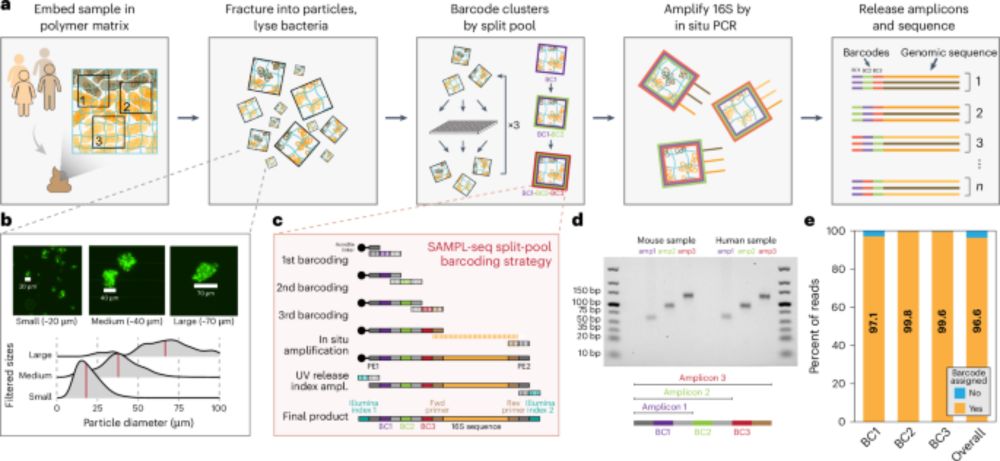
SAMPL-seq reveals micron-scale spatial hubs in the human gut microbiome - Nature Microbiology
Split-And-pool Metagenomic Plot-sampling sequencing (SAMPL-seq) can be applied to complex microbial communities to reveal spatial co-localization of microbes at the micron scale.
Happy to share our new paper in @naturemicrobiol.bsky.social on mapping spatial relationships of the human gut microbiome. We identified distinct spatial hubs between gut bacteria that reflect sub-community assemblies at the micron-scale. Led by Miles Richardson & co.
www.nature.com/articles/s41...
03.02.2025 14:46 — 👍 89 🔁 36 💬 0 📌 0
I hear ya! I mean more on the “we” rather than “I” personally in some of these sentiments. Again, this is not meant as a substitute for things we do or aught to do already but something that can augment. Also, just an idea to stimulate conversations.
16.12.2024 04:24 — 👍 0 🔁 0 💬 0 📌 0
I like your ideas, just trying to think of ways that I can donate my “time” to help specific labs/areas that might even be outside of my immediate area of expertise. Seems like having a universal currency for science might have some uses. It’s obviously imperfect, but maybe could refine the ideas.
15.12.2024 19:46 — 👍 0 🔁 0 💬 1 📌 0
I think the idea is that you can compensate others with your credits/tokens like pay for reagents (service contracts on equipment!!). There are lots of misc expenses in running a lab that add up!
15.12.2024 19:40 — 👍 0 🔁 0 💬 0 📌 0
May not push the incentives to a point where people will review more, but at least extend the effort of the review to benefit not only the authors (or the journals) but broaden impact to others through the peer review process. It’s like volunteering time and using earned credits to give to others.
15.12.2024 19:39 — 👍 0 🔁 0 💬 1 📌 0
The problem is that people don’t have time to review. It’s not clear how you easily “fact check” papers in bio (unless there are massive flaws). Critiques of strengths & weaknesses are still useful. Most people aren’t volunteering time to review biorxiv papers. Maybe this system can be an incentive.
30.11.2024 22:21 — 👍 0 🔁 0 💬 1 📌 0
Credits used back to invest in science community rather than on direct personal benefit.
30.11.2024 06:15 — 👍 2 🔁 0 💬 1 📌 0
I recognize that this idea is still underdeveloped but wanted to start share it here. Would love to hear feedback and find people willing to help do something like this! Thanks for reading! (9/9)
30.11.2024 05:14 — 👍 8 🔁 0 💬 3 📌 0
I can imagine this framework on a blockchain to track credits or tokens and you can have people invest in buying the tokens directly to further add to this community (basically a donation). Lots of ways to grow this organically. The economics need to be figured out in more detail. (8/9)
30.11.2024 05:14 — 👍 2 🔁 0 💬 2 📌 0
Importantly you can DONATE these credits to other labs or entities. What if there’s a cool project from another lab that I want to support? I can donate my science credits to them so it can advance the science. You basically converted ↔️ PR time/effort into communal goods for sharing w/ others. (7/9)
30.11.2024 05:14 — 👍 7 🔁 0 💬 1 📌 0
The SOLUTION:💡What if we gave reviewers some type of “science credits” after peer review. Sci-credits can be exchanged for all kinds of science related activities. For example: publication fees, society/conference fees, discount on purchasing reagents or repair cost of equipment. (6/9)
30.11.2024 05:14 — 👍 12 🔁 0 💬 3 📌 2
News from the Rubin Lab 🧬🧫 We conduct DNA-level editing of microbial communities for understanding and control. Managed by lab members.
@innovativegenomics.bsky.social @ucberkeleyofficial.bsky.social
🔗 therubinlab.org
Postdoc in Mooney Lab @Harvard/Wyss | PhD @Columbia | Bacterial Therapy, Engineered Living Materials, Synthetic Biology
harimotolab.com
passionate about discovering ways to leverage early life microbiome/ immune development to prevent autoimmune disease, pediatric infectious disease physician and immunologist.
Postdoc @Columbia in NYC • BWFPDEP scholar • Archaea fanatic • @SFSU @JohnsHopkins alumni • Moving from extremophiles ➡️ gut, using CRISPR as my guide✂️🧬🦠
physician-scientist, author, editor
https://www.scripps.edu/faculty/topol/
Ground Truths https://erictopol.substack.com
SUPER AGERS https://www.simonandschuster.com/books/Super-Agers/Eric-Topol/9781668067666
Assistant Professor of Applied Physics @Stanford. Theoretical biophysics, evolutionary dynamics & microbial evolution. https://bgoodlab.github.io/
The Vilcek Foundation raises awareness of immigrant contributions in the United States and fosters appreciation of the arts, sciences, and humanities.
Learn more at https://vilcek.org
Postdoctoral Fellow @TypasLab @EMBLHeidelberg
Interested in ecology and evolution of microbial communities, microbiome, culturomics, experimental (co)evolution, microbiome modulation
🦠🧫🔬🧪💊💩🧬
https://scholar.google.com/citations?user=dhZfxqMAA
Evolutionary biochemist and protein complex enthusiast at the University of Marburg.
Senior editor @science.org. Molecular biology, #DNA, #RNA, #gene regulation, #epigenetics, nuclear biology, #chromatin biology, 3D #genome, #synbio, #CRISPR and gene editing, other bacterial immune systems, and #AI in all these
Chemical Biologist at Penn Medicine. Burslemlab.com
Confused Brit in the US. He/him
7th Annual Canadian Synthetic Biology and Bioengineering conference
University of Toronto, June 1-3, 2025
https://synbio7.vercel.app/
science reporter covering biomedical research at Nature | proudly Ukrainian 🇺🇦
maxkozlov.com
signal: mkozlov.01
(my views don't reflect those of my employer, springer nature)
Biochemist 🧪| Empowering Life Scientists and Science Communicators to Achieve Their Career Goals | UCLA PhD + NIH Alum | Pharma R&D @GSK + @AstraZeneca 💊 | #SciComm Director ✍️| Career Coach | Star Trek | Sherlock Holmes | 🎹 | https://larrymillerphd.com
Passoniate microbiome research; www.clavel.ukaachen.de
Bioengineering Postdoc @ Stanford
a CRISPR company - www.SNIPRBIOME.com
Cell Press partners with scientists across all disciplines to publish and share work that will inspire future directions in research. #ScienceThatInspires
Assistant Professor of Bioengineering at Stanford











