This is an important letter! We often do ‘omics analysis and each of the 1000’s of analytes measured has its own power calculation which is impossible to predict.
21.02.2026 20:31 — 👍 3 🔁 1 💬 0 📌 0
So cool and so many research questions from these data! @savitski-lab.bsky.social -lab.bsky.social
07.02.2026 13:42 — 👍 1 🔁 0 💬 0 📌 0
Big thanks to @unimelbcmr.bsky.social and many others!
05.02.2026 18:36 — 👍 0 🔁 0 💬 0 📌 0
This looks fun and super easy! Thanks! Do you have to run all existing QC runs with new runs simultaneously in a single search? Or can you add new QC search results of day only a few files to existing searches?
28.01.2026 09:35 — 👍 2 🔁 0 💬 1 📌 0
Thanks @nickescott.bsky.social @unimelbcmr.bsky.social and many others
06.01.2026 20:23 — 👍 0 🔁 0 💬 0 📌 0
Yes but the issue is false positives with phosphorylated Ser/Thr. A cocktail of phosphatases combined with mild beta-elimination may reduce this… www.sciencedirect.com/science/arti...
27.11.2025 19:10 — 👍 1 🔁 0 💬 1 📌 0
It’s interesting that the 8600 doesn’t appear to be marketed with single cell capabilities compared to other vendors. I wonder why SCIEX didn’t at least have a note in this space?
26.09.2025 19:57 — 👍 1 🔁 0 💬 1 📌 0
Two weeks to go before the abstract deadline (July 18th)! Looking forward to seeing you all there!
04.07.2025 13:42 — 👍 6 🔁 1 💬 0 📌 0
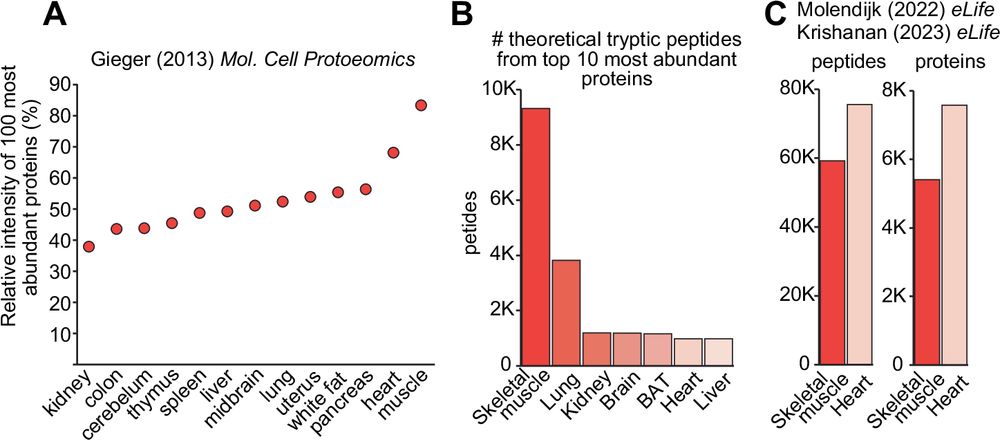
Skeletal muscle proteomics: considerations and opportunities
npj Metabolic Health and Disease - Skeletal muscle proteomics: considerations and opportunities
Are you interested in #proteomics and #skeletalmuscle? It can be a real pain to get deep reproducible coverage. Check out our review with @adeshmukh.bsky.social We analyse some considerations including current limitations and how to potentially overcome them #myoblue @unimelbcmr.bsky.social
03.07.2025 18:17 — 👍 19 🔁 4 💬 0 📌 0

👩💻 We’re hiring! Lê Cao Lab at Uni Melbourne @mig-unimelb.bsky.social needs an R dev to power the next-gen of mixOmics 🚀
Love #RStats, #Bioconductor & multi-omics? Help expand mixOmics, run workshops & publish cutting-edge methods.
Apply: unimelb.wd105.myworkdayjobs.com/en-US/UoM_Ex...
23.06.2025 22:20 — 👍 11 🔁 12 💬 0 📌 2
LinkedIn
This link will take you to a page that’s not on LinkedIn
🚨 Has PRIDE helped your research?
Take 15 mins to tell funders why open data matters!
📊 Fill out the EMBL-EBI 2025 survey 👉
www.surveymonkey.com/r/QGFMBH8?ch...
Your feedback helps keep PRIDE open, FAIR & impactful.
🙏 Please share!
#FAIR #OpenData #Proteomics #MassSpectrometry #PRIDE
18.06.2025 06:02 — 👍 10 🔁 12 💬 0 📌 0
This sounds fantastic and can’t wait to try it on some new peptidomics data!
06.06.2025 10:44 — 👍 0 🔁 0 💬 1 📌 0
Congratulations Atul and team! Amazing body of work.
27.05.2025 19:54 — 👍 1 🔁 0 💬 0 📌 0
Yes probably redundant, but there are considerations before pushing back. Do you have lysate remaining or would you have to repeat experiments? How many other ‘battles’ are there in the comments? If there are bigger fights and you have lysate than might be worth a ‘quick’ WB to appease the gods.
14.05.2025 19:07 — 👍 0 🔁 0 💬 1 📌 0
Thanks for all the support @unimelbcmr.bsky.social!!
11.05.2025 20:09 — 👍 0 🔁 0 💬 0 📌 0
Big thanks to all our collaborators for making this wok possible. @unimelbcmr.bsky.social @craigagoodman1.bsky.social and many others…
11.05.2025 20:04 — 👍 0 🔁 0 💬 0 📌 0

Why do we care? Because we have previously shown that UFC1 levels are negatively correlated with lean mass in various mouse strains, and knockdown of UFC1/UFMylation can increase muscle function. Maybe myosin UFMylation is a key link? elifesciences.org/articles/82951
11.05.2025 20:04 — 👍 0 🔁 0 💬 1 📌 0

We quantified UFMylation in the above human biopsies revealing several increased UFMylation sites including Myosins! Additional mapping of sites on Myosins reveal they are extensively modified, and in silico modelling suggest sites adjacent to ATP binding plays a functional role.
11.05.2025 20:04 — 👍 0 🔁 0 💬 1 📌 0

What did we find?... A whole bunch of cool substrates including UFMylation of Myosins which we validate following knockdown of UFC1 (the E2 ligase) in vivo combined with TMT-16plex labelling and LC-MS/MS…
11.05.2025 20:04 — 👍 2 🔁 0 💬 1 📌 0

In collaboration with CST, we developed antibodies that recognise "remnant ValGly" left on UFM1-conjugates following tryptic digestion. Similar to the remnant ubiquitin "diGly" approach published by www.cell.com/molecular-ce...
11.05.2025 20:04 — 👍 0 🔁 0 💬 1 📌 0

However, check out this anti-UFM1 blot from control muscle biopsies and people with ALS... A big band at 30kDa (probably RPL26) but clearly more to discover including big heavy bands >250kDa!
11.05.2025 20:04 — 👍 0 🔁 0 💬 1 📌 0
UFMylation is a ubiquitin like modification of UFM1 applied to substrate lysine residues. <15 substrates have been identified (a nice summary: www.cell.com/molecular-ce.... RPL26 has been most extensively characterised and suggested to be the major substrate…
11.05.2025 20:04 — 👍 1 🔁 0 💬 1 📌 0
Very nice indeed. In this example does LLOQ=LLOD?
08.05.2025 11:24 — 👍 2 🔁 0 💬 1 📌 0

Exciting news: We have released #FragPipe 23, and it's one of our biggest updates ever. Windows installer, support for TMT on Astral and timsTOF, TMT35, PTM site reports for DIA, improved Astral data handling in #MSFragger, improved diaTracer for diaPASEF data, better Skyline integration, and more!
05.05.2025 04:06 — 👍 80 🔁 15 💬 4 📌 1
The grant agencies in Australia seem to specifically announce outcomes on Friday afternoons. I think the idea is so you can cool off over the weekend. However, better than Monday morning!
11.04.2025 18:05 — 👍 1 🔁 0 💬 1 📌 0
Yes, this is where things are interesting. All databases produce similar number of total peptides. Large db’s identify more novel peptides missing in canonical db but the larger search space and requirement for higher score at 1%FDR means peptides get filtered out.
22.03.2025 18:22 — 👍 1 🔁 0 💬 0 📌 0
Lab content for researchers 🧫 | Join 1,000+ researchers getting weekly lab hacks with our newsletter (it’s free) 👉 wildtypeone.substack.com/about
Evolutionary molecular biologist/biochemist | Postdoc @ UQ, Brisbane | former PhD @ EMBL, Heidelberg | Sponges, other non-Bilaterians and proteomics
Postdoc in Lehtiö group at SciLifeLabs/Karalinsa. Canadian abroad 🇨🇦. Proteomics and PTMs.
Geneticist, Skeletal Muscle, Drug development, Gene therapies. Non-coding RNA, and Zebrafish Aficionado. Lover of all things muscle. All posts are my own. Instagram @thealexanderlab
Lab website: https://www.uab.edu/medicine/peds/research/division-researc
Assistant Professor at Leiden University. Covalent inhibitors and chemoproteomics for antibiotics. Views my own. he/him/his. https://orcid.org/0000-0001-5420-4824
Biological Mass Spectrometry
Proteomics at Scale
Scientist at Seer
Opinions independent of employer
Biological Mass Spectrometry enthusiast, trying to make sense of cellular hierarchies and heterogeneity
Lead Bioinformatician in the Stroud Lab @UniMelb Bio21 Institute | Computational Mass Spec Multi-Omics | Chair of ABACBS Diversity, Inclusion and Accessibility Committee
Professor in Exercise Physiology
Work: https://tinyurl.com/4z884z2p
https://scholar.google.com/citations?hl=en&user=sLZykm8AAAAJ
PostDoc in Olsen Group and Arachnid wll-being executive manager. 🕷️🕸️ Working on Spatial Proteomics and extracellular vesicles. 🤗
Proteins | Sugars | Yeast | Beer | Mass Spectrometry | Professor in Biochemistry UQ. CW chickens, fungi, cats, pups.
Science, glycobiology, mass spectrometry, coffee. Assoc. professor @unimelb. Former ARC future fellow. he/his
Mass spectrometrist and laser collector. Posting mass spectrometry articles from journal RSS feeds. https://kermitmurray.com/
🔬 We research and develop treatments for cancer, diseases of development and ageing, immune disorders and infections.
https://www.wehi.edu.au
Cell & Developmental Biology. Vascular Biology. Lymphatics. Zebrafish.
Associate Director of Laboratory Research at Peter Mac Cancer Centre. Professor at The University of Melbourne.
A small band of nerds using genes, light, and maths to understand the zebrafish brain. Preaching diversity and good vibes in science.
🇦🇺🇺🇸🏴🇸🇨🇬🇷🇨🇦🇬🇧🇮🇳🇪🇹🇩🇰🇨🇳🇧🇷🏳️🌈
UniMelbourne, Australia.
Lab website: tinyurl.com/bdfey9bk
Scientist in the School of Biotechnology and Biomolecular Sciences (BABS) at UNSW Sydney studying gene regulation and transcription factors in adipose tissue-resident immune cells and red blood cells
All things muscle - synthesis, breakdown, damage and regeneration.
Postdoc at @ismcopenhagen.bsky.social