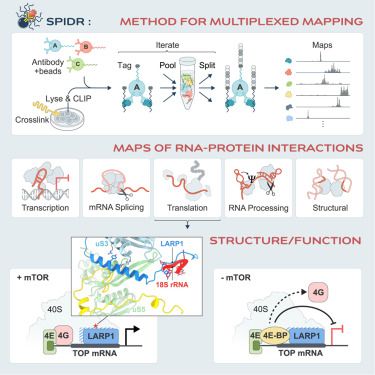
SPIDR enables multiplexed mapping of RNA-protein interactions and uncovers a mechanism for selective translational suppression upon cell stress @cellcellpress.bsky.social @mitchguttman.bsky.social
www.cell.com/cell/fulltex...
@mitchguttman.bsky.social
Molecular biologist interested in non-coding RNAs, nuclear organization, and gene regulation. Professor at Caltech

SPIDR enables multiplexed mapping of RNA-protein interactions and uncovers a mechanism for selective translational suppression upon cell stress @cellcellpress.bsky.social @mitchguttman.bsky.social
www.cell.com/cell/fulltex...
This work was led by co-first authors Jimmy Guo and Erica Wolin with support from amazing teams from our lab @caltech.edu and the Jovanovic lab @columbiauniversity.bsky.social and @jbquerido.bsky.social lab with financial support from NHGRI, NSF, @genometdcc.bsky.social.
26.07.2025 19:15 — 👍 3 🔁 0 💬 0 📌 0
We used SPIDR to identify mTOR-dependent changes and observed that 4EBP1 showed a dramatic increase in binding upon mTOR inhibition specifically at mRNAs containing a TOP-motif, suggesting a new model for how translational repression is selectively achieved upon mTOR inhibition.
26.07.2025 19:15 — 👍 2 🔁 0 💬 1 📌 0
We identified an interaction between LARP1 and 18S rRNA located within the mRNA channel entry site on the 40S small ribosomal subunit and @jbquerido.bsky.social resolved this structure at 2.8 Å using single-particle cryo-EM.
26.07.2025 19:15 — 👍 2 🔁 0 💬 1 📌 0
Single nucleotide binding maps generated by SPIDR can map known RNP structures at atomic resolution and identify novel components within RNP structures.
26.07.2025 19:15 — 👍 1 🔁 0 💬 1 📌 0
We show that SPIDR generates high quality data across a diverse range of RBPs, including transcription, splicing, translation, and miRNA biogenesis, all within a single experiment.
26.07.2025 19:15 — 👍 1 🔁 0 💬 1 📌 0
SPIDR uses a dramatically simplified split-and-pool based strategy to increase the throughput of CLIP by two orders of magnitude. SPIDR enables the rapid generation of consortium-level datasets within any molecular biology lab without the need for specialized training or equipment.
26.07.2025 19:15 — 👍 1 🔁 0 💬 1 📌 0
Many proteins bind RNA, yet we still don’t know what RNAs most bind because methods map one RBP at a time. In @cp-cell.bsky.social, with the Jovanovic lab, we describe SPIDR – a method for mapping the RNA binding sites of dozens of RBPs in a single experiment. www.sciencedirect.com/science/arti...
26.07.2025 19:15 — 👍 99 🔁 37 💬 1 📌 2This work was led by co-first authors Drew Perez and Isabel Goronzy and our amazing team @caltech.edu, and funding from NHGRI.
27.11.2024 04:13 — 👍 11 🔁 0 💬 0 📌 0
By enabling the generation of consortium-level datasets within any molecular biology lab, ChIP-DIP facilitates a fundamental transition from ‘reference-maps’ to context-specific maps and represents a transformative new tool for dissecting cell-type specific gene regulation.
27.11.2024 04:13 — 👍 11 🔁 0 💬 1 📌 0
We used ChIP-DIP to explore quantitative combinations of histone modifications that define distinct classes of regulatory elements and integrated these signatures with regulatory factor binding to identify their functional activity.
27.11.2024 04:13 — 👍 5 🔁 0 💬 1 📌 0
We used ChIP-DIP to measure temporal chromatin dynamics in primary mouse dendritic cells following stimulation and correlate these with transcriptional changes. For this, we mapped multiple time points within a single experiment, multiplexing across both proteins and samples.
27.11.2024 04:13 — 👍 5 🔁 0 💬 1 📌 0
ChIP DIP generates highly accurate maps for ALL classes of DNA-associated proteins, including histone modifications, chromatin regulators, transcription factors, and RNA polymerases.
27.11.2024 04:13 — 👍 5 🔁 0 💬 1 📌 0
ChIP-DIP uses a simple antibody-labeling strategy followed by split-and-pool barcoding to multiplex DNA-protein mapping. This enables the rapid generation of consortium-level datasets within any molecular biology lab without the need for specialized training or equipment.
27.11.2024 04:13 — 👍 8 🔁 0 💬 1 📌 0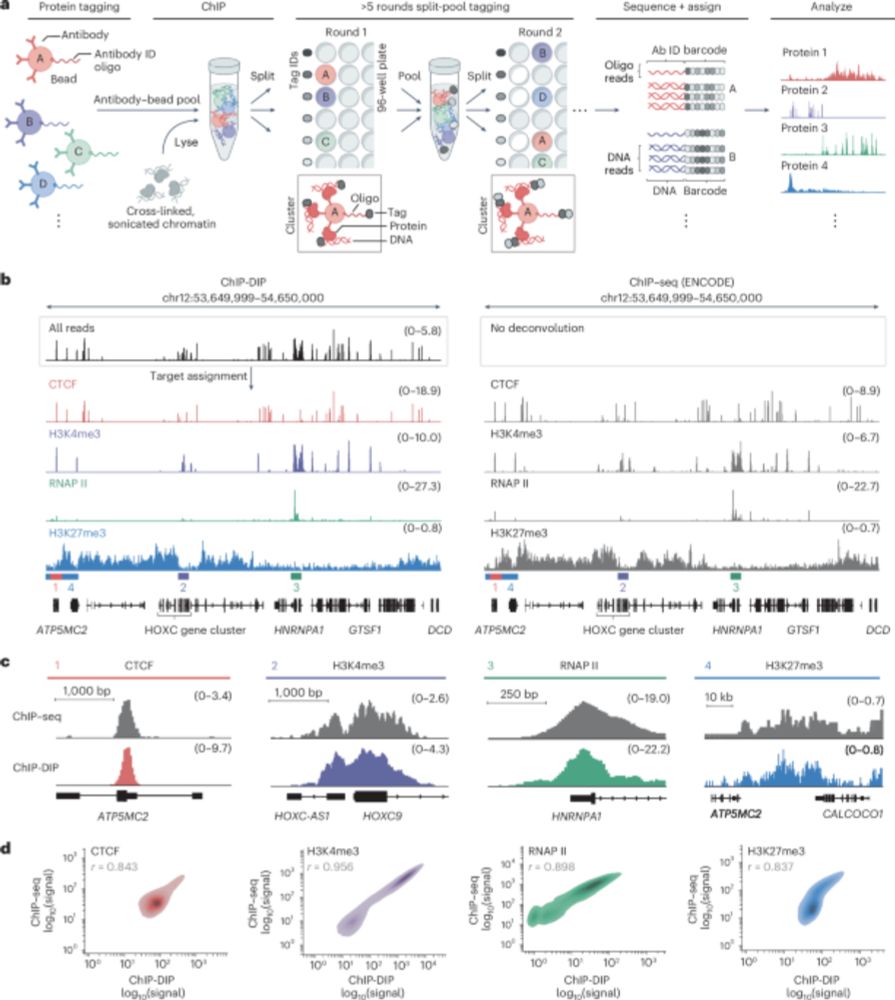
Gene regulation involves thousands of proteins that bind DNA, yet comprehensively mapping these is challenging. Our paper in Nature Genetics describes ChIP-DIP, a method for genome-wide mapping of hundreds of DNA-protein interactions in a single experiment.
www.nature.com/articles/s41...
Great thread on our paper from Jan👇
22.11.2024 18:39 — 👍 13 🔁 5 💬 0 📌 0
A report made counterintuitive claims that PRC2 binds
→ more RNA than PTBP1 & hnRNPU
→ RNAs that do not exist in the cell
We found a simple explanation: the authors ignore most RNA in the sample, leading to inflated background & misleading conclusions.
www.biorxiv.org/content/10.1...
Leonid Mirny and I wrote this for all interested in chromosomes: "The chromosome folding problem and how cells solve it"
www.cell.com/action/showP...