Good point on the learning part. Backprop seems difficult to implement using molecules, currently contemplating other means, e.g. the exhaustive search strategy you mentioned, or Hebbian learning
16.12.2024 05:54 — 👍 0 🔁 0 💬 1 📌 0
I am so glad the algorithm has brought me to this gem
15.12.2024 07:55 — 👍 0 🔁 0 💬 0 📌 0
Thank you, Preetham!
15.12.2024 07:41 — 👍 0 🔁 0 💬 0 📌 0
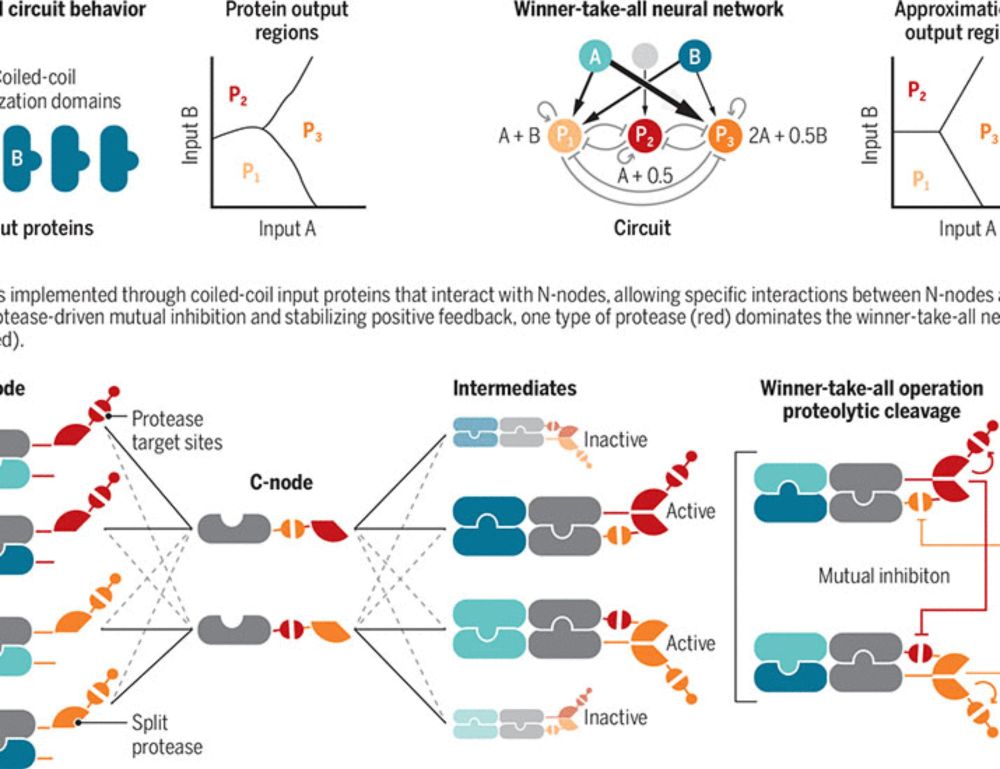
Bringing neural networks to life
A synthetic protein-based winner-take-all neural network controls cell fate decisions
Finally, thanks to Katie Galloway and Christopher Johnstone for this thoughtful perspective. We are indeed excited about the "learning/training" aspect of the Perceptein network! www.science.org/doi/10.1126/...
13.12.2024 13:46 — 👍 1 🔁 0 💬 0 📌 0
It was tremendous fun brainstorming with
@elowitzlab.bsky.social
in the early days of this project, and collaborating with all co-authors on this paper. A nice cover art made by the talented Ehmad Chehre:
13.12.2024 13:45 — 👍 3 🔁 0 💬 1 📌 0
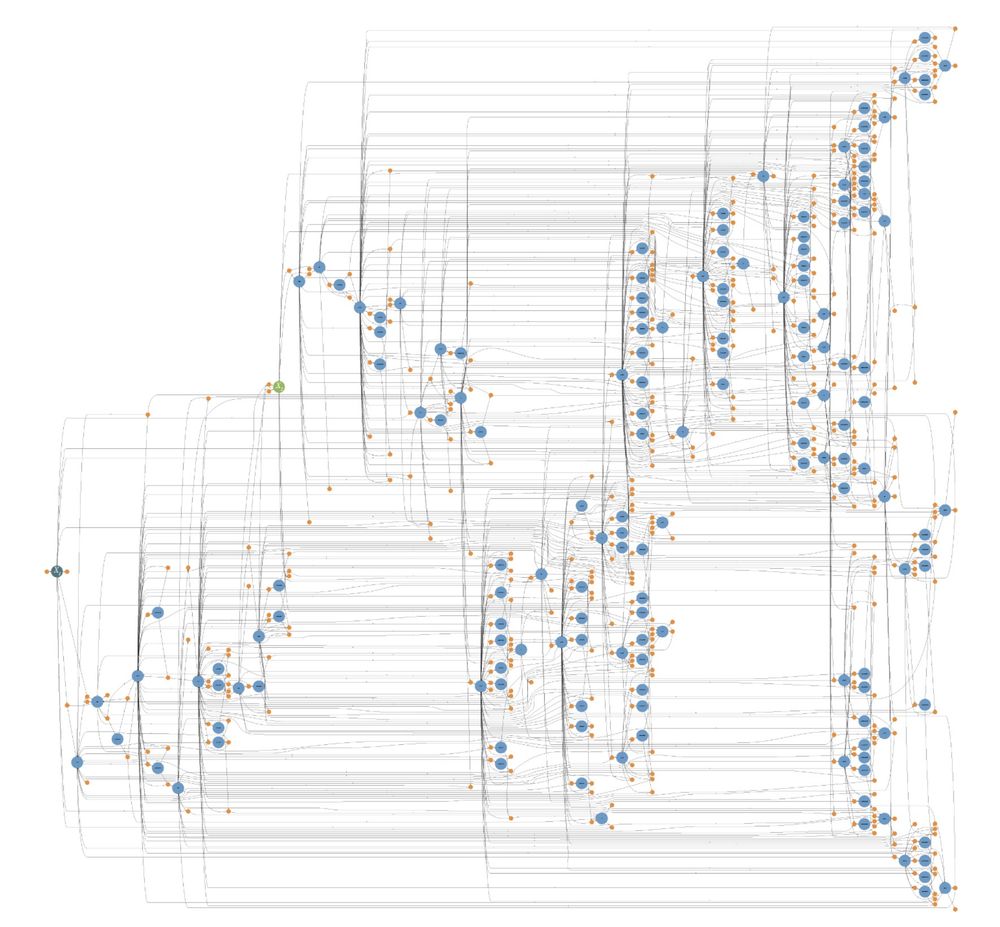
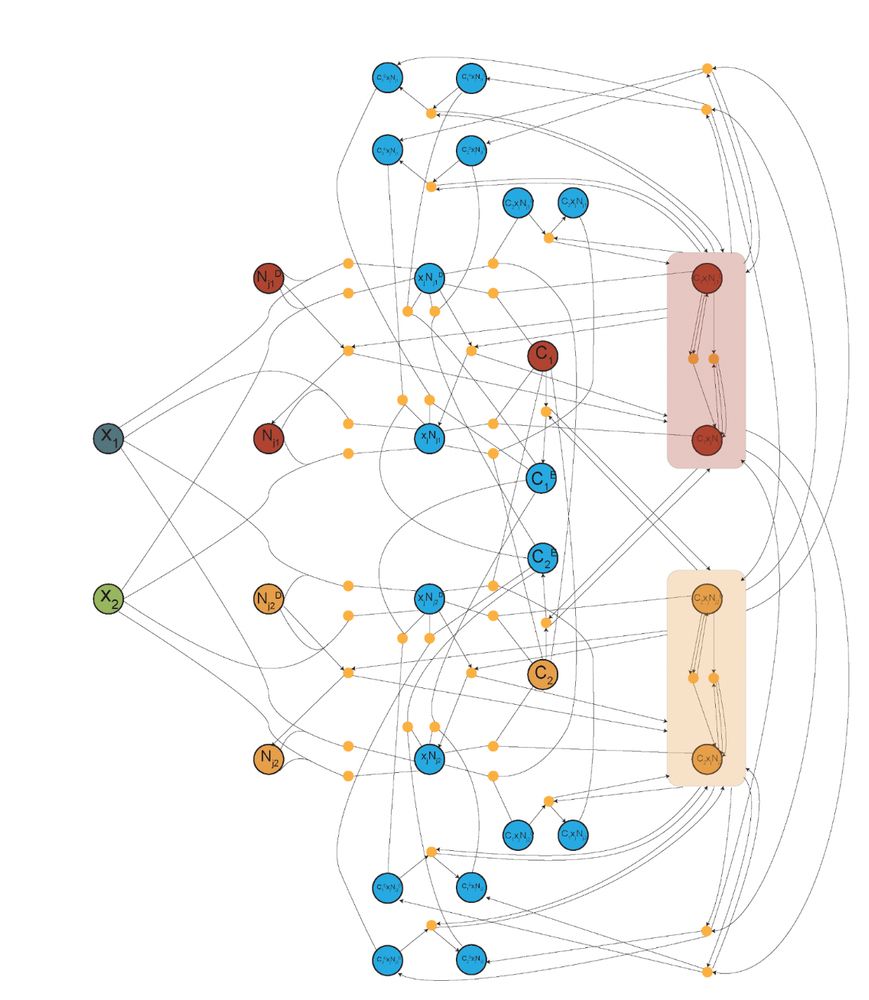
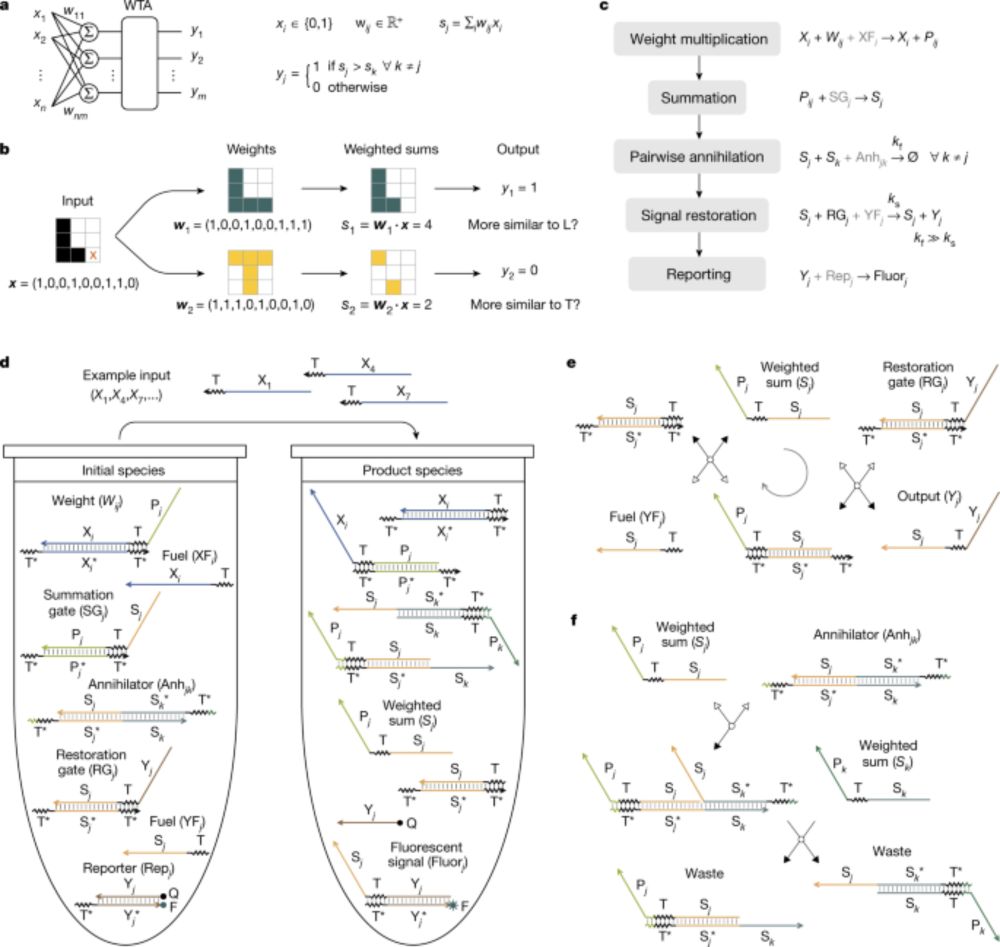
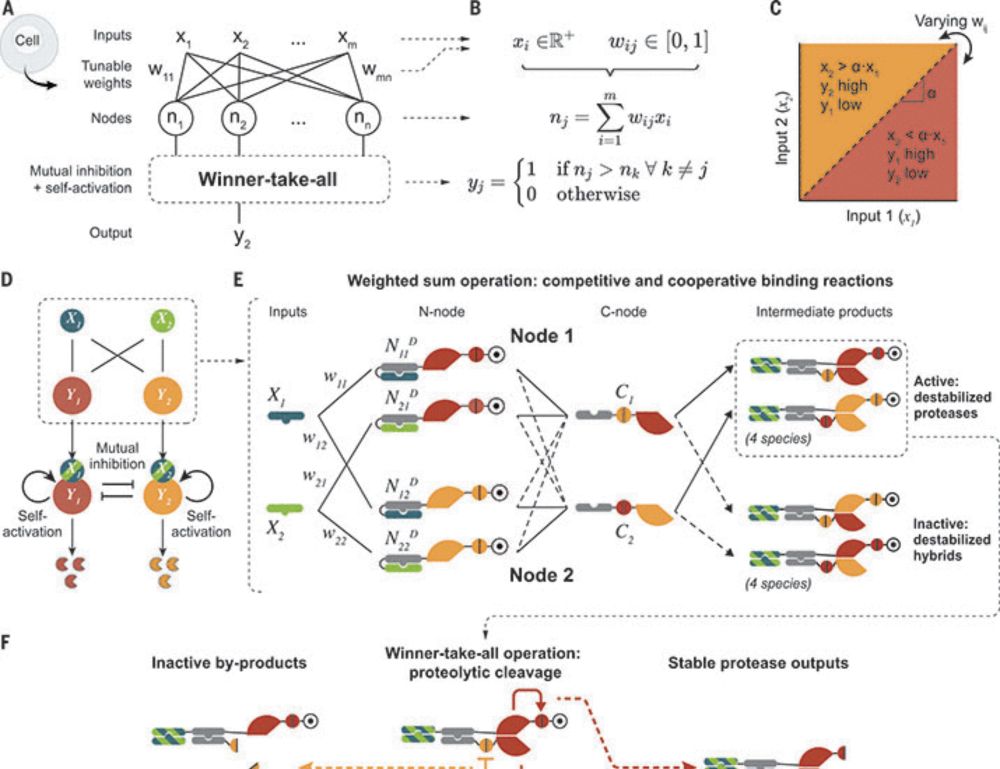
3) cleaned up the chemical reaction network diagram (from left to right). Here, each circle is a unique (left) or a group of (right) protein species, and each orange dot represents one (left) or a group of (right) chemical reactions
13.12.2024 13:44 — 👍 1 🔁 0 💬 1 📌 0
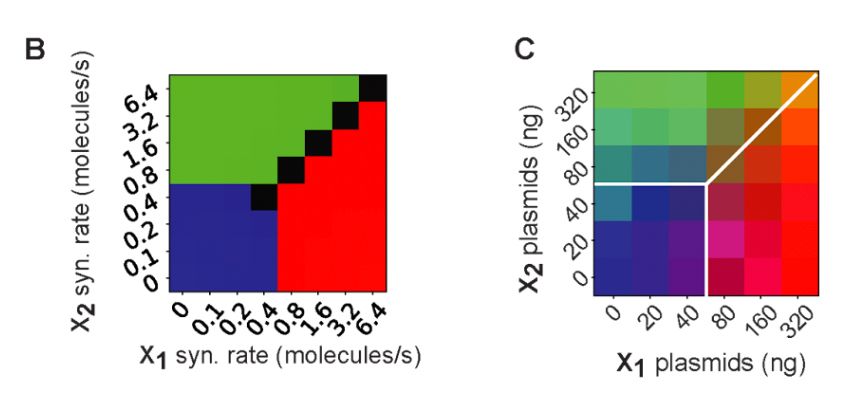
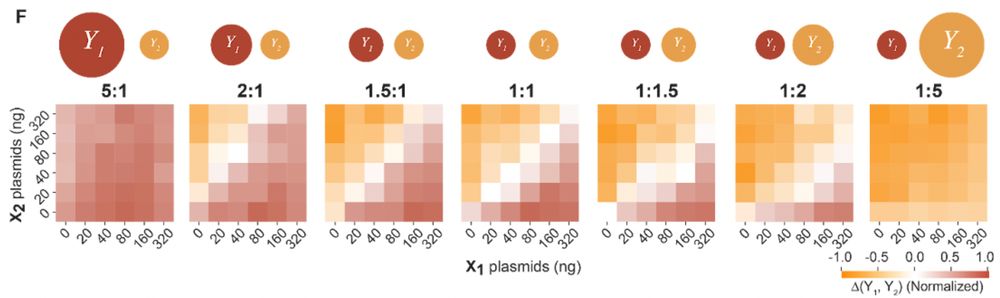
2) scaled up the neural network to be 2-input and 3-output, showcasing the scalability of the Perceptein architecture (left, simulation; right, experiments)
13.12.2024 13:44 — 👍 0 🔁 0 💬 1 📌 0
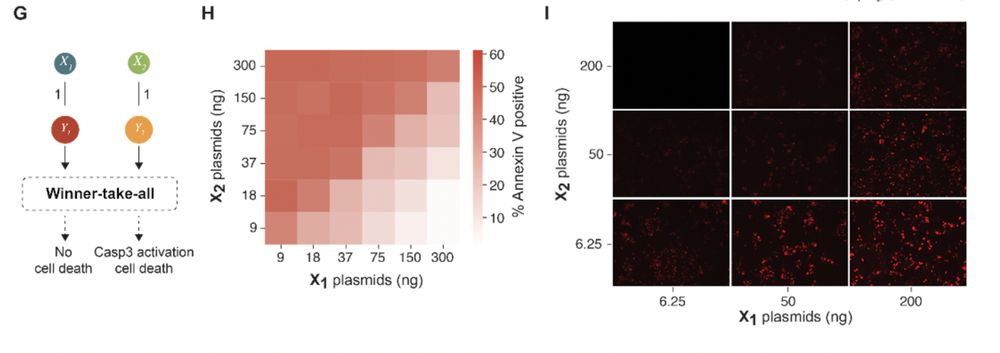
Since our last preprint, we have
1) redirected the classification outcome to cell death, demonstrating the interfacability of protein circuits (thanks to Shiyu Xia)
13.12.2024 13:43 — 👍 1 🔁 0 💬 1 📌 0
As shown in our preprint a while ago, we took a small step further and created a protein circuit, made of de novo designed protein heterodimers and engineered split viral proteases, that carries out weights-tunable winner-take-all neural network computation in mammalian cells.
13.12.2024 13:42 — 👍 1 🔁 0 💬 1 📌 0
Executive Editor at Science
The Alon lab explores design principles of biological circuits, focusing on systems medicine, aging, and healthspan.
Assistant Professor, Columbia University | Mobile elements, Structural biology and Genome engineering | http://thawanilab.org
Single cell systems and synthetic biology lab at Northwestern University and Chan Zuckerberg Biohub Chicago.
https://www.goyallab.org/
Focused on developing molecular technologies to control cell fate with precise spatiotemporal resolution. Recovering computational geneticist, ex-Altos, ex-eGenesis. Caltech PhD.
We study quantitative microbial cell physiology.
https://jun.ucsd.edu
“If you want to go fast, go alone. If you want to go far, go together.”
-African Proverb
Schmidt AI in Science Postdoctoral Fellow at U. Chicago
theoretical & computational soft matter, biophysics, machine learning
jordanshivers.github.io
Computational structural biologist @Fox Chase Cancer Center. Gay, out & proud since 1986. I support transgender & nonbinary people. Living with CLL/SLL. Video about my LGBTQ+ life in science: https://tinyurl.com/45srzjdw. Views my own, not my employer's.
News from the King Lab kinglab.ipd.uw.edu.
Part of @uwmedicine.bsky.social and @uwproteindesign.bsky.social.
Some assembly required.
Assistant Professor @ Westlake University. PhD @ MPI-IS. Focusing on democratizing human-centric digitization.
A biochemistry PhD candidate at Caltech in the Elowitz Lab studying systems and synthetic biology! First-gen and Cal State LA alum! He/Him
Postdoc in Ahmad (Mo) Khalil's lab at Harvard University / Boston University. Synthetic multicellularity | Understanding and engineering multicellularity via synthetic biology and experimental evolution.
@DamonRunyon Postdoc Fellow in Tim Springer lab @harvardmed and @BostonChildrens | Organoids, Integrins, Antibodies, CryoEM | @Cornell & @IISERPune alumnus | He/Him
#STEM+Arts education. I am Tamuka, Ph.D aka Dr. Muk, a Biochemist IRL & I started #hiphopbiochemistry 🇿🇼🐝🦄🏳🌈 he/him | My opinions are my own linktree.com/steamulater
Postdoc in the Zhang lab @ Broad Institute. Formerly in the Elowitz lab @ Caltech.
Née Ke-Huan Chow.
I ❤️ SynBio and cats.
HHMI fellow of the Damon Runyon Cancer Research Foundation in Elowitz lab at Caltech | PhD in Ovi Chaudhuri lab at Stanford
Curious scientist leading a research team mainly working on organelle biology and biotechnology: we study -and often “torture”- chloroplasts to understand how they do their magic ;)
https://www.oeaw.ac.at/gmi/research/research-groups/silvia-ramundo
Lorenzo Di Michele's research group at @cebcambridge.bsky.social University of Cambridge (and also a bit at Imperial College). Working on DNA/RNA nanotechnology and Synthetic Cells
PhD @dimichelelab1.bsky.social working on 🧬 and synthetic cells @cebcambridge.bsky.social | Chemistry graduate at ICL | interested in all things Outreach 🧪🔬
postdoc at the weizmann institute (bar-ziv lab) - tinkering with gene expression - bottom-up synthetic biology