
Our annual PhD call is closing at the end of this week on 30 November. If you're interested in carrying out world-class scientific research in Barcelona, you still have a few days left to submit your application! www.crg.eu/en/content/t...
25.11.2025 07:43 — 👍 13 🔁 19 💬 0 📌 3

Thinking about doing a PhD with us? We're running an online Q&A session just for you! Hear practical advice directly from our experts in the Training and Academic Office and register here: events.teams.microsoft.com/event/26a7dc...
04.11.2025 10:34 — 👍 2 🔁 2 💬 0 📌 0

"Meet the companies": a chance to boost your career - El·lipse
In this yearly event, PRBB residents get to know professional opportunities beyond academia through informal conversations with companies on a wide range of areas in the biomedical and biotech sector.
All set for "Meet the Companies"❗️
A unique gathering connects PRBB residents with a dozen leading companies in diagnostics, gene editing, venture capital, patent law, scientific management, communication and citizen science.
🗓️4 November, 15h
ℹ️
02.11.2025 21:01 — 👍 1 🔁 1 💬 0 📌 0

Are you looking for a PhD? Join us in Barcelona! You'll dive into a community of >100 PhD students from 30 countries exploring the frontiers of biology. You can also join an online workshop on 6 November (15:00 CET) to learn how to find the right lab for you.
More info: www.crg.eu/en/content/t...
23.10.2025 07:55 — 👍 25 🔁 29 💬 1 📌 8

Bernardo Rodriguez-Martín EvoMG Program, CRG
Next was Bernardo Rodriguez-Martín ( @bernardo-rodriguez.bsky.social ), from the EvoMG Program and @crg.eu, on highly mutagenic (hot) L1 polymorphisms in cancer
12.10.2025 09:09 — 👍 1 🔁 1 💬 1 📌 0
We are excited that the Earth BioGenome Project, a global network of scientists including those from the Sanger Institute, has mapped the second phase of its ambitious plan to sequence all 1.67 million known species on Earth by 2035. 👏
www.sanger.ac.uk/news_item/bi...
05.09.2025 08:34 — 👍 27 🔁 7 💬 0 📌 0

28 | Caroline Bartman and the flash(cards) of inspiration
Podcast Episode · Night Science · 02/13/2023 · 28m
A great thing about working in a lab is being involved in lab meetings and witnessing everyone's projects take shape. A single project may seem to go slow but all projects together is impressive. Caroline Bartman discussed this on the podcast:
podcasts.apple.com/us/podcast/2... @cbartman.bsky.social
19.08.2025 03:00 — 👍 25 🔁 6 💬 0 📌 0
Thanks to the huge contributions from all the co-authors of this large effort, who are listed in the main thread, and to the reviewers, whose thoughtful feedback served to greatly improve the manuscript since its initial submission 🙏
31.07.2025 07:32 — 👍 0 🔁 0 💬 0 📌 0

GitHub - REPBIO-LAB/SVAN: Structural Variants ANnotator (SVAN)
Structural Variants ANnotator (SVAN). Contribute to REPBIO-LAB/SVAN development by creating an account on GitHub.
These insights were largely possible thanks to SVAN (github.com/REPBIO-LAB/S...). While many tools call SVs, none systematically analyse their sequences to infer mechanisms. SVAN fills this gap by taking a VCF from your favourite SV caller and adding annotations of SV classes and features [9/9]
31.07.2025 07:32 — 👍 0 🔁 0 💬 1 📌 0

We identified 10 HERVK, 7 of them not previously reported, and 28 solo-long terminal repeats (LTRs) [7/9]
31.07.2025 07:32 — 👍 0 🔁 0 💬 1 📌 0

Transduction tracing uncovers locus-specific patterns in source L1 and SVA elements. While most L1s mediate only 3′ transductions, some SVAs show 5′ or 3′ specificity. A striking exception: one highly active L1 uses promoter hijacking to drive 5′ transductions. [6/9]
31.07.2025 07:32 — 👍 0 🔁 0 💬 1 📌 0

We observed large sequence diversity among L1 and SVA inserts, with dozens of structural conformations shaped by their integration mechanisms and transductions. SVA elements show a broad size range, largely driven by variability in their internal hexameric and VNTR repeats. [5/9]
31.07.2025 07:32 — 👍 0 🔁 0 💬 1 📌 0

Our new study scales up to the population level: we sequenced 1,019 human genomes with ONT, resolving the sequence of 23,212 Alu, 4,851 L1, and 3,239 SVA insertions, a gain of +20% (Alu), +166% (L1), and +179% (SVA) over short-read analysis of the same samples. [4/9]
31.07.2025 07:32 — 👍 0 🔁 0 💬 1 📌 0

Recurrent inversion polymorphisms in humans associate with genetic instability and genomic disorders
Large-scale analysis of haplotype-resolved inversions in human genomes unveils recurrent
inversion polymorphisms and their disease relevance.
Refreshment of Chapter 2: “Twin priming”.
Microhomology patterns observed at 5´ inversion and truncation breakpoint junctions for 1.271 L1 insertions point to MMEJ. We also found three templated insertions, suggestive of polymerase theta. [3/9]
www.cell.com/cell/fulltex...
31.07.2025 07:32 — 👍 0 🔁 0 💬 1 📌 0

Haplotype-resolved diverse human genomes and integrated analysis of structural variation
Human genetic variation is elucidated from de novo assembly of 32 genomes selected as representatives of the 1000 Genomes Project.
Refreshment of Chapter 1: “Hot L1s, age and coding potential”.
MEI analysis on 32 haplotype-resolved human genomes. Older L1s have higher allele frequencies, lower ORF preservation and are therefore less active, but eternal youth can occur [2/9]
www.science.org/doi/10.1126/...
31.07.2025 07:32 — 👍 1 🔁 0 💬 1 📌 0
Sequence-resolved mobile element sagas, Chapter 3: “MEI at population scale”
This work closes our trilogy on MEI research advances enabled by long-read sequencing. Bonus tutorial about MEI of our structural variation study of 1KGP samples resequenced using @nanoporetech.com [1/9]
31.07.2025 07:32 — 👍 0 🔁 0 💬 1 📌 0
Congratulations to all awardees with a #PID2024!! 🥳 extremely happy and thrilled to share our project proposal (L1-CANPREDICT) got funded. Keep tuned about the research to come from REPBIO at @crg.eu
30.07.2025 10:31 — 👍 2 🔁 0 💬 0 📌 0

A man is smiling. Colourful houses of Gamla Stan are visible in the background.
Travel broadens the mind – an excellent example of that is EMBL ARISE Fellow Thomas Weber, who expanded his data science skills during his secondment at @scilifelab.se.
👇 Read the full story about his journey and the impact of his ARISE Fellowship.
www.embl.org/news/people-...
29.07.2025 12:41 — 👍 9 🔁 2 💬 0 📌 0
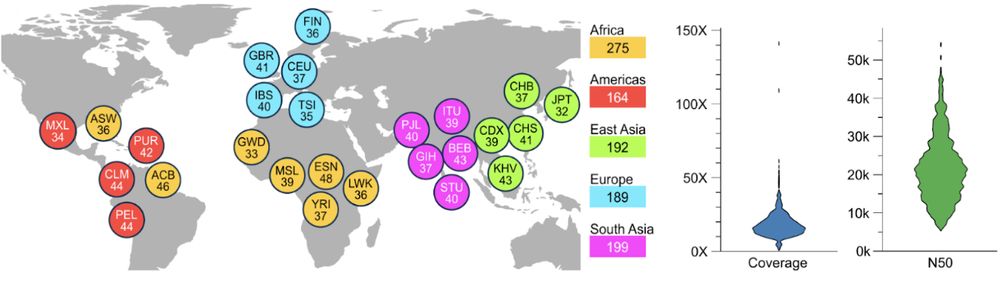
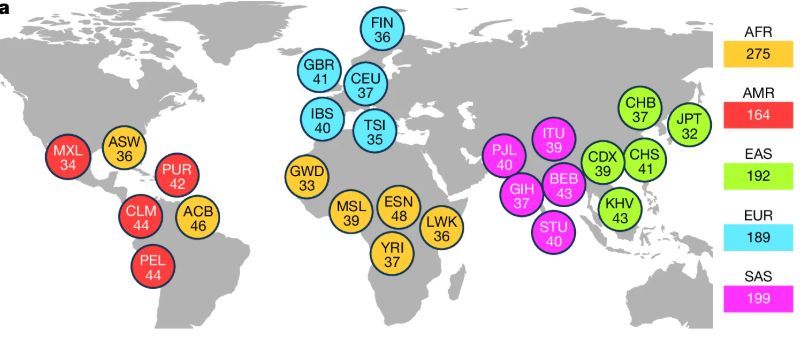
[1/8] *New Open-Access Long Read Resource*. We sequenced 1,019 genomes from the 1000 Genomes Project sample cohort using @nanoporetech.com long-read sequencing (LRS) to median 17x coverage. Publication at go.nature.com/4ffPb8f.
@hhu.de @crg.eu @embl.org @impvienna.bsky.social
24.07.2025 09:26 — 👍 43 🔁 21 💬 1 📌 2

EMBL researchers and their collaborators have provided exciting new insights into human genetic variation by building upon the 1000 Genomes Project dataset.
The two studies constitute what may be the most complete overview of the human genome to date.
www.embl.org/news/science...
23.07.2025 15:19 — 👍 28 🔁 8 💬 1 📌 0
The first, co-authored by @bernardo-rodriguez.bsky.social, discovered more than 167,000 structural variants across the 1,019 individuals, doubling the known amount of structural variation in the human pangenome. Most variants were rare, which will help accelerate the diagnosis of rare diseases.
23.07.2025 15:50 — 👍 2 🔁 1 💬 1 📌 0
Evolution of antibiotic resistance. Ramón y Cajal Fellow at Universidad Autónoma de Madrid. https://amrevolution.es/
A platform for life sciences. Publications, research protocols, news, events, jobs and more. Sign up at https://www.lifescience.net.
We are the largest biomedical science hub in the south of Europe, by the beach in Barcelona.
Recerca biomèdica amb vistes |
@researchmar.bsky.social @crg.eu @melisupf.bsky.social @embl.org @isglobal.org @ibe-barcelona.bsky.social @fpmaragall.bsky.social
The goal of the Canadian BioGenome Project is to produce high-quality reference genomes 🧬 for all Canadian species 🌎
Sequencing Canada's Biodiversity 🌿🦋🐍🧬🐢🦌🐸🌳🐿🐙🦈🦦
Learn more: https://linktr.ee/canadianbiogenome
Neuroscientist, Centro de Biología Molecular Severo Ochoa (CSIC-UAM)
MSCA fellow at @crg.eu w/M. Dias and @jonnyfrazer.bsky.social
> Biological Physics | Proteins | Comp Bio | ML
https://scholar.google.com/citations?user=n55NtEsAAAAJ&hl=en
Bioinformatics Scientist / Next Generation Sequencing, Single Cell and Spatial Biology, Next Generation Proteomics, Liquid Biopsy, SynBio, AI/ML in biotech // http://albertvilella.substack.com
Director of Language & Genetics at Max Planck Institute, Nijmegen.
Tracing the complex connections between genes, brains, speech & language.
Website: https://www.mpi.nl/people/fisher-simon-e
ORCID: https://orcid.org/0000-0002-3132-1996
Aging and cancer stem cell heterogeneity - ICREA research professor - Quantitative Stem Cell Dynamics lab
at IRB Barcelona 🇦🇷🇪🇺🇺🇸 fraticellilab.com
Account of the Saez-Rodriguez lab at EMBL-EBI and Heidelberg University. We integrate #omics data with mechanistic molecular knowledge into #opensource #ML methods
Website: https://saezlab.org/
GitHub: https://github.com/saezlab/
Group leader @crg.eu | blood, single cell, synthetic genomics
SNSF and EMBO Postdoctoral fellow at EMBL | PhD in Dev Bio from University of Zurich | Utrecht University alumni | climbing | cycling | piano
Computational Biologist in Singapore. Nocturnal baker and maker of caffeinated beverages.
My lab at Stanford studies human population genetics and complex traits.
Professor of Computer Science @ JHU. https://www.langmead-lab.org/ https://www.youtube.com/BenLangmead
Responsible for instigating some of humanity’s most high-impact discoveries.
Visit us at https://science.ucsc.edu/ 🐋 🔭 🧬
(Software | Data | Knowledge Graph) Engineer.
Working in the Personalized Health Informatics group
at SIB Swiss Institute of Bioinformatics
Thoughts and views are my own.
The GREGoR Consortium (Genomics Research to Elucidate the Genetics of Rare diseases) seeks to develop and apply approaches to discover the cause of currently unexplained rare genetic disorders.
https://gregorconsortium.org/
Professor, University of Michigan. Scholar of genomes, reader of books (sci-fi, fantasy), player of games (board, video), and camper of forests.





















