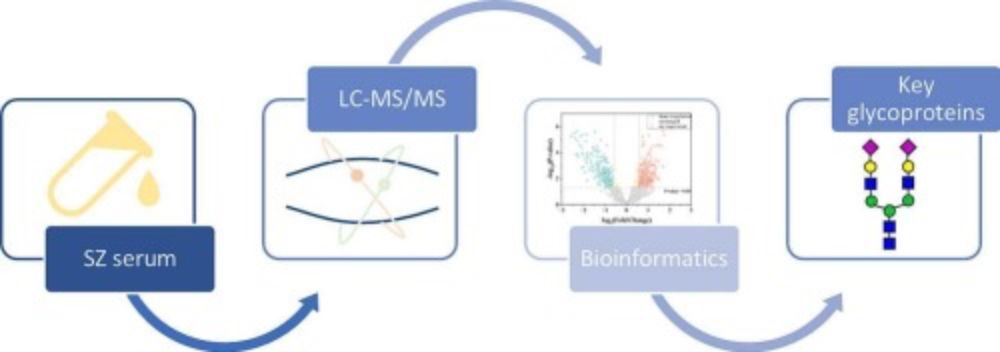
Serum N-glycoproteomics characterization of differential N-glycosylation in schizophrenia #JProteomics #MassSpec www.sciencedirect.com/science/arti...
20.03.2025 19:54 — 👍 0 🔁 1 💬 0 📌 0@promiselab.bsky.social
Proteome-level Microbial Systems Ecology (ProMiSE) Lab

Serum N-glycoproteomics characterization of differential N-glycosylation in schizophrenia #JProteomics #MassSpec www.sciencedirect.com/science/arti...
20.03.2025 19:54 — 👍 0 🔁 1 💬 0 📌 0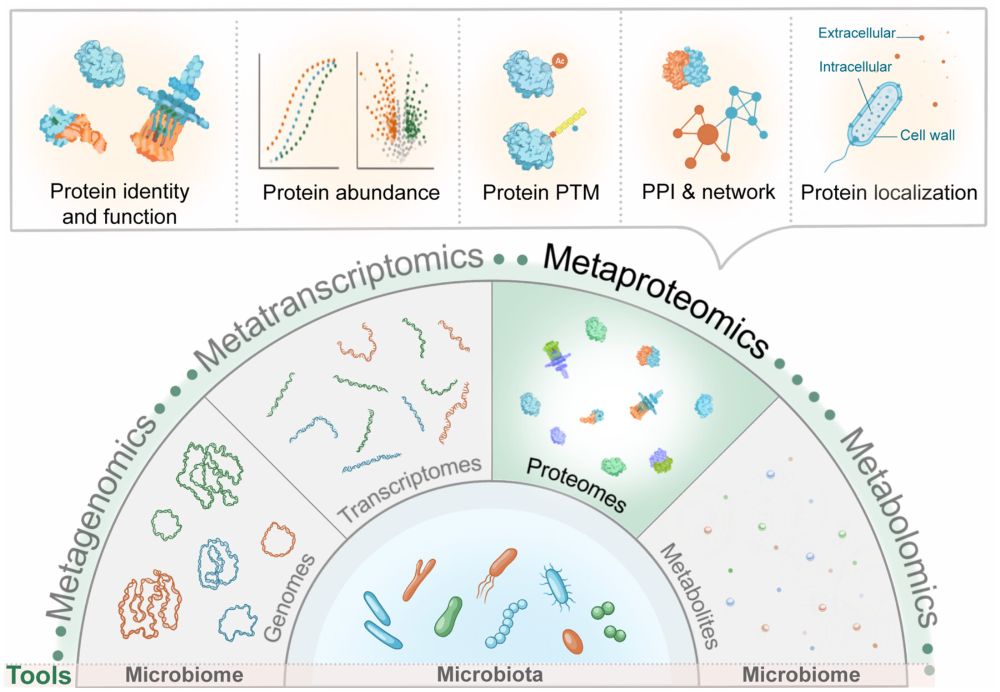
The microbiologist's guide to metaproteomics
onlinelibrary.wiley.com/doi/full/10.... #Bioinformatics #microbiome #metaproteomics #proteomics #MS
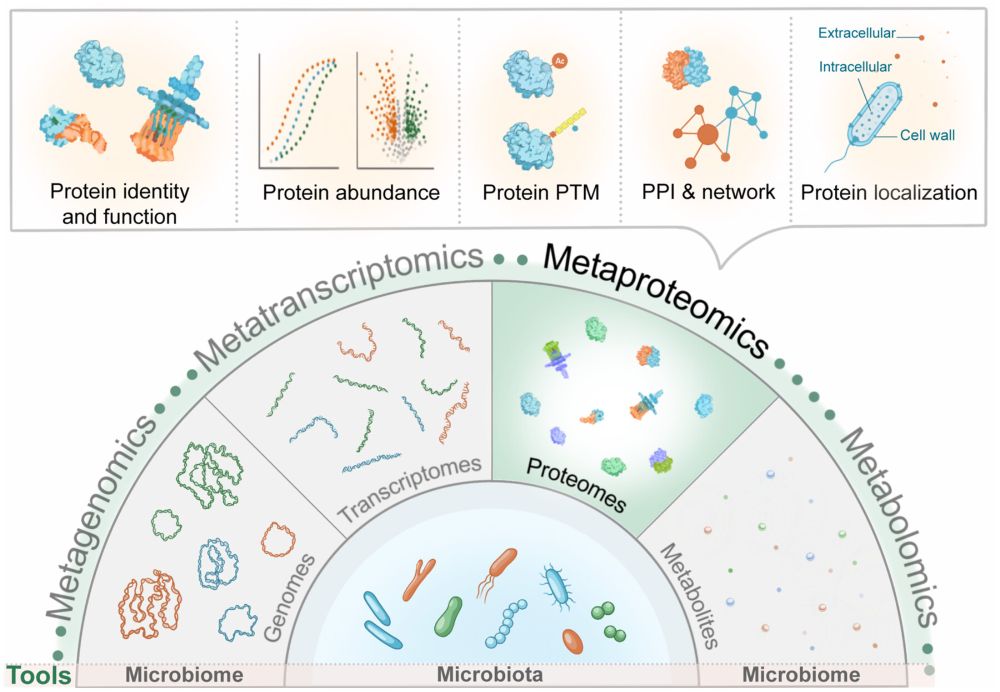
Are you new to #metaproteomics but don't know where to start? Then this guide, written by experts of the Metaproteomics Initiative, is for you! onlinelibrary.wiley.com/doi/full/10....
08.05.2025 03:47 — 👍 6 🔁 1 💬 0 📌 0
🚀 Interested in the interactions of xenobiotics with the gut microbiome? Join us @quadraminstitute.bsky.social to apply precision microbiome assays to xenobiotics. Apply by 28 May 2025. 👉 quadram.ac.uk/vacancies/gr...
#microbiome #PrecisionNutrition #metaproteomics #metabolics #metagenomics
Compared to conventional methods, OSaMPle increased bacterial peptide identifications by 3.2x and protein identifications by 1.7x. Using OSaMPle, we analyzed mouth rinse samples from IBD patients and found significant shifts in bacterial protein expression. 3/3 #Metaproteomics
28.04.2025 09:04 — 👍 0 🔁 0 💬 0 📌 0We developed OSaMPle (Optimized Salivary MetaProteomic sample analysis workflow) to enrich bacterial cells from saliva and mouth rinse, reducing host contamination and enabling deeper oral metaproteomics. 2/3
28.04.2025 09:04 — 👍 0 🔁 1 💬 1 📌 0The oral microbiome is linked to various inflammatory conditions, including IBD. However, analyzing functional changes at the protein level has been challenging due to host protein interference. 🧵 1/3
28.04.2025 09:04 — 👍 2 🔁 1 💬 1 📌 0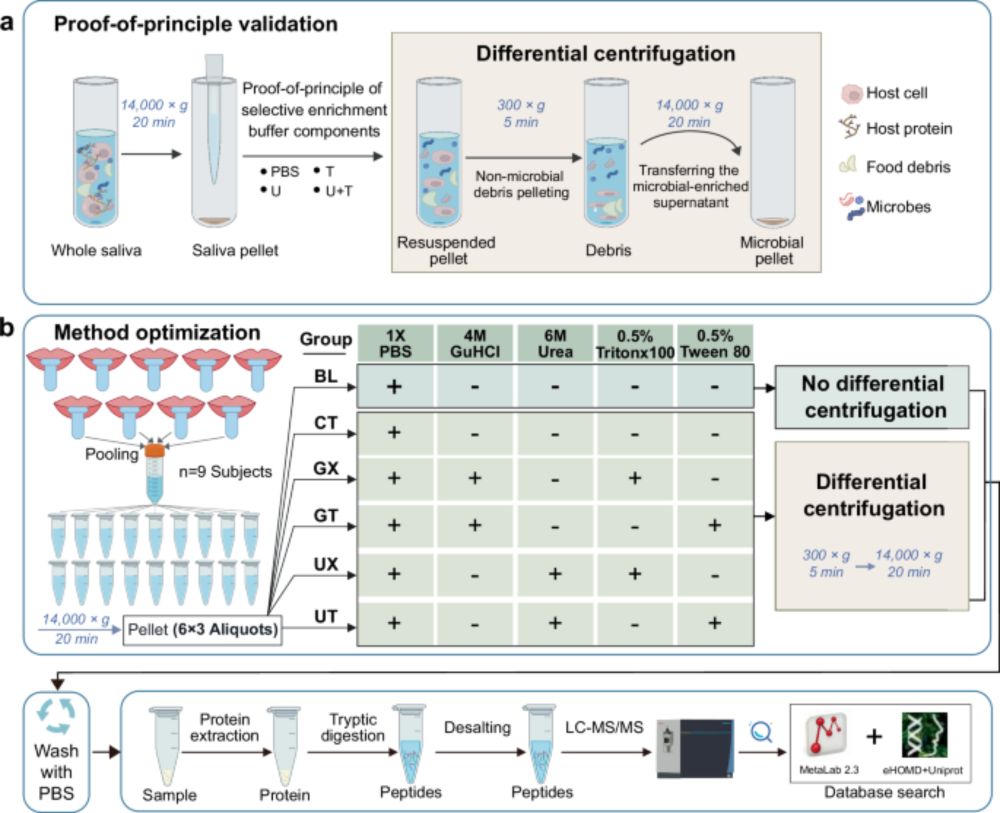
I couldn’t be prouder—this is the work of the very first master's student that I've supervised. Salivary metaproteomics is tricky due to host/debris interference. Yuan et al. developed a microbial cell enrichment step and achieved cleaner data with better insights. www.nature.com/articles/s41...
24.04.2025 12:01 — 👍 1 🔁 0 💬 0 📌 1
Metaproteomics deserves a dedicated β-diversity metric:
microbiolly.wordpress.com/2025/03/19/p...

🚨 We mapped 4.6M #microbial protein responses to 312 compounds, uncovering 3 distinct functional gut #microbiome states and revealing how neuropharma triggers deep shifts (lower functional redundancy, higher #AMR proteins).
www.biorxiv.org/content/10.1...
microbiomejournal.biomedcentral.com/articles/10....
12.02.2025 02:44 — 👍 0 🔁 0 💬 0 📌 0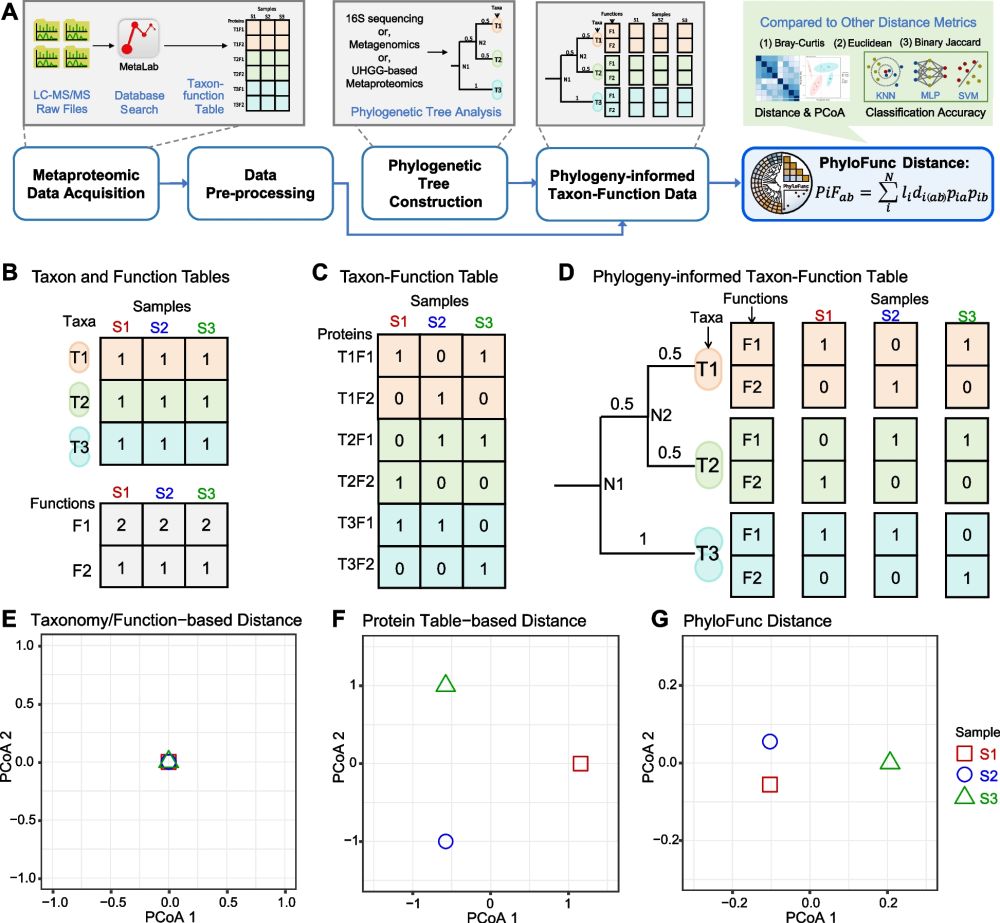
🦠 In #metaproteomics, proteins are often treated as independent features, ignoring their ecological relationships.
🌱 We developed PhyloFunc, a beta diversity metric that integrates phylogeny into functional distance calculations, offering a more ecologically meaningful way to compare microbiomes.

🎉 Excited to share this amazing Metaproteomics Initiative community effort aims to make metaproteomics accessible and easy to use for microbiome researchers. Grateful for this unforgettable journey of collaboration! 🌍🔬 #Metaproteomics #Collaboration #Science chemrxiv.org/engage/chemr...
12.01.2025 06:49 — 👍 8 🔁 3 💬 0 📌 0Thank you for sharing Manuel!
12.01.2025 06:25 — 👍 0 🔁 0 💬 0 📌 0
If you want to get started with #metaproteomics , this guide is a good starting point. This was an amazing metaproteomics community effort to get all the key information together led by Tim Van Den Bossche and Leyuan Li
chemrxiv.org/engage/chemr...