Come join us in Basel! @biozentrum.unibas.ch is an amazing place to start your group.
29.09.2025 18:35 — 👍 17 🔁 5 💬 0 📌 0
Please let us know what you think. And much agreed on Bach and Vermeer (maybe I should give Proust another try).
29.09.2025 13:35 — 👍 0 🔁 0 💬 0 📌 0
Are you excited to pursue your own research interests and contribute to our group’s experimental research in the quantitative biology of microbes? We have an opening for a potentially permanent position as research associate.
Please repost and notify potential candidates!
18.09.2025 09:26 — 👍 3 🔁 12 💬 0 📌 0
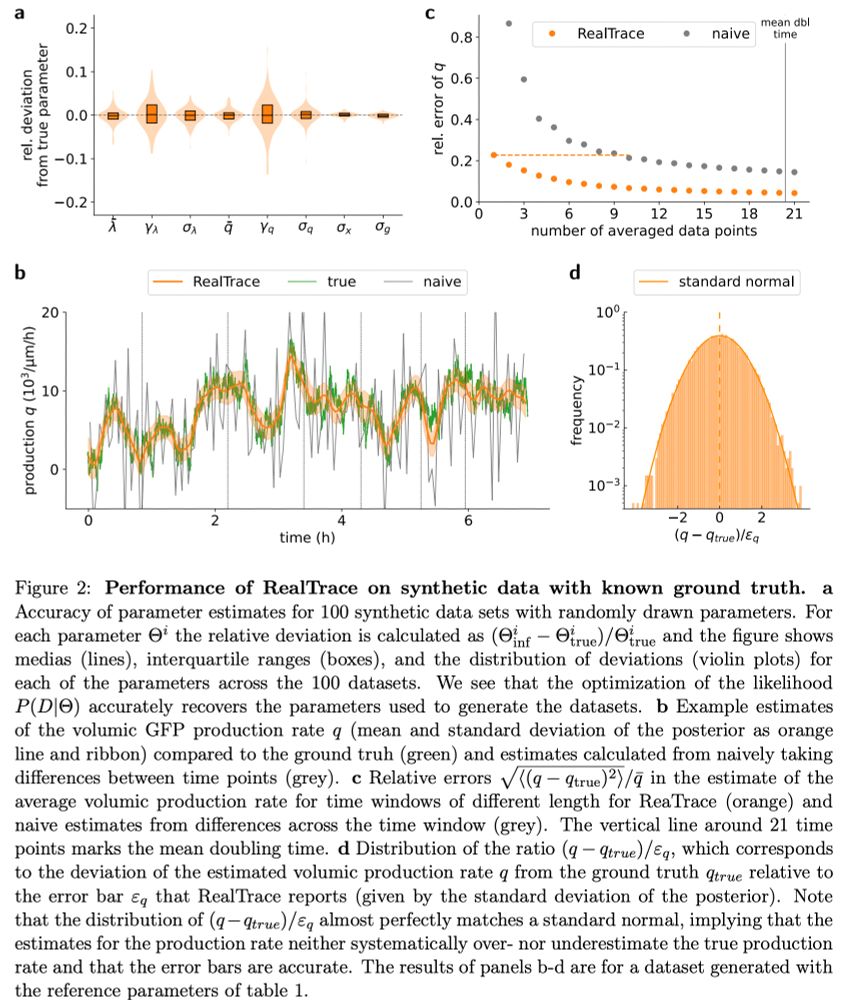
and includes accurate error-bars on all estimates.
We believe RealTrace can dramatically enhance the power of time-lapse microscopy data by allowing identification and quantification of far more subtle features in the dynamics than is possible with simple data smoothing approaches. n/n
17.09.2025 14:18 — 👍 0 🔁 0 💬 0 📌 0
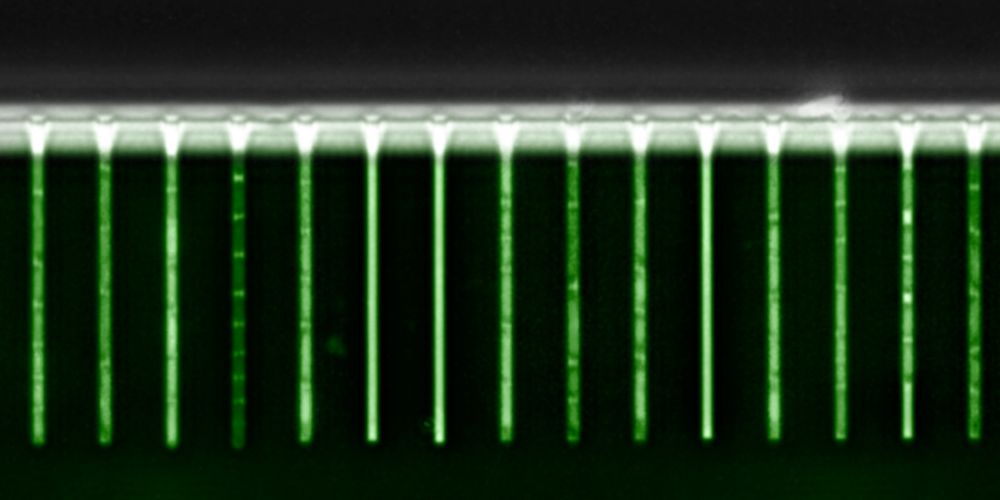
Surprisingly, protein production rates adapt to their new steady-states far more quickly and with only a minimal transient.
RealTrace can be applied to essentially any time-lapse fluorescence microscopy data and gives accurate estimates of instantaneous growth and volumic GFP production rates, 10/n
17.09.2025 14:18 — 👍 1 🔁 0 💬 1 📌 0

Finally, upon a sudden `downshift' in nutrients, we see very reproducible time-dependent responses across single cells. All cells go almost into growth arrest, then overshoot in growth rate, only to relax to a final growth rate over a time scale of 3 cell cycles. 9/n
17.09.2025 14:18 — 👍 0 🔁 0 💬 1 📌 0

minimum early in the cell cycle and a maximum late in the cell cycle. In contrast, protein production rates show more complex patterns that vary across conditions and across promoters, even across constitutive promoters!
As far as we are aware, no current models can explain these patterns. 8/n
17.09.2025 14:18 — 👍 0 🔁 0 💬 1 📌 0
time, and in slower growth conditions, both the amplitude and duration of these fluctuations increases.
Second, both growth rate and volumic protein production vary systematically across the cell cycle. Interestingly, the variation in growth rate is the same in all conditions, with a 7/n
17.09.2025 14:18 — 👍 0 🔁 0 💬 1 📌 0

We use data from E. coli cells carrying fluorescent reporters of constitutive and ribosomal genes, growing in a microfluidic device in different conditions, to highlight the kind of subtle patterns RealTrace can uncover.
First, instantaneous growth rates vary substantially across cells and 6/n
17.09.2025 14:18 — 👍 0 🔁 0 💬 1 📌 0

RealTrace is very generally applicable, which we demonstrate by applications to data from E. coli cells, mouse embryonic stem cell nuclei (data from the lab of @davidsuter.bsky.social), and entire C. elegans larvae (data from the lab of @betowbin.bsky.social). 5/n
17.09.2025 14:18 — 👍 0 🔁 0 💬 1 📌 0

are correlated on short time scales, all measurement errors are independent. We implemented this idea into a rigorous Bayesian procedure that uses maximum entropy process priors and recursively approximates the non-linear dynamics over short time intervals. 4/n
17.09.2025 14:18 — 👍 2 🔁 0 💬 1 📌 0
We can't fit the data to particular curves because we typically don't know what form the dynamics will take, and local smoothing systematically distorts the true dynamics by replacing it with a time average.
RealTrace solves this challenge using only one assumption: while biological changes 3/n
17.09.2025 14:18 — 👍 0 🔁 0 💬 1 📌 0
Tracking the behavior of single cells using fluorescence time-lapse microscopy is becoming increasingly popular but there is a fundamental challenge in analyzing such data: as biological changes are small on short time scales, much of the true dynamics is hidden under measurement noise. 2/n
17.09.2025 14:18 — 👍 0 🔁 0 💬 1 📌 0
We're looking for highly motivated candidates for PhD positions in the physics of living systems.
Our group uses theoretical physics to understand how cells collectively self-organize.
If you're interested, get in touch & check out the Biozentrum PhD Fellowship program, deadline October 12th!
15.09.2025 10:04 — 👍 14 🔁 9 💬 0 📌 1

Join us for an exciting #BiozentrumDiscovery lecture!
Prof. Tami Lieberman from @mit.edu will speak about the dynamic selective landscape of human microbiomes. Her lab studies microbial evolution in real time, with a focus on mutations occurring within individual human microbiomes.
08.09.2025 09:13 — 👍 20 🔁 6 💬 1 📌 0
Anybody taking bets on how obvious the cherry-picking/p-hacking is going to turn out to be to obtain this supposed association?
I'm personally going with: mind-blowingly glaringly obvious. Like 'My mouth is wide open but I still cannot breathe at the stupidity of it' obvious. But hey, who knows?!
06.09.2025 16:47 — 👍 3 🔁 0 💬 1 📌 0

Multi-omic assessment of mRNA translation dynamics in liver cancer cell lines
Scientific Data - Multi-omic assessment of mRNA translation dynamics in liver cancer cell lines
Hey there, #mRNAtranslation buffs! Our latest multi-faceted dataset from liver cancer cell lines is out in #ScientificData: RNA-seq, ribosome run-off/ribo-seq, polysome-seq, pSILAC. rdcu.be/eC86H. Great team effort spearheaded by Asier Gonzalez Sevine and Niels Schlusser. Happy data crunching!
30.08.2025 11:40 — 👍 8 🔁 1 💬 0 📌 0
Are you saying that is bad? I already KNOW I'm interested in your lab.. I just used the LLM to help me express it.
28.08.2025 19:44 — 👍 1 🔁 0 💬 0 📌 0

Cellular memory: The clever strategy cells use to move through narrow environments
In wound healing, immune response, and cancer metastasis, cells migrate through the body – often squeezing through narrow spaces. Researchers led by Prof. David Brückner at the Biozentrum discovered t...
🧪How do cells navigate the body? David Brückner @davidbrueckner.bsky.social & researchers @umons.bsky.social discovered that cells retain a “memory” of their past shapes, allowing them migrate more efficiently–crucial for immune response & metastasis. @unibas.ch @natphys.nature.com ow.ly/SSzX50WLg3t
25.08.2025 10:04 — 👍 36 🔁 12 💬 2 📌 1
I agree many of the unprocessed foods were poorly presented. But I think one can’t conclude from that that it must taste bland. I remember after first coming to the US having birthday pie. Extravagantly colorful and shiny. But it tasted much more bland than the pies I got in Europe.
06.08.2025 04:34 — 👍 1 🔁 0 💬 0 📌 0
I feel the thread is vastly exaggerating how much better the processed food looks. In some cases, just based on looks, I would prefer the unprocessed one.
Maybe this study is evidence of imprinting of US citizens on what ‘tasty’ food is supposed to look like.
06.08.2025 02:16 — 👍 3 🔁 0 💬 1 📌 0
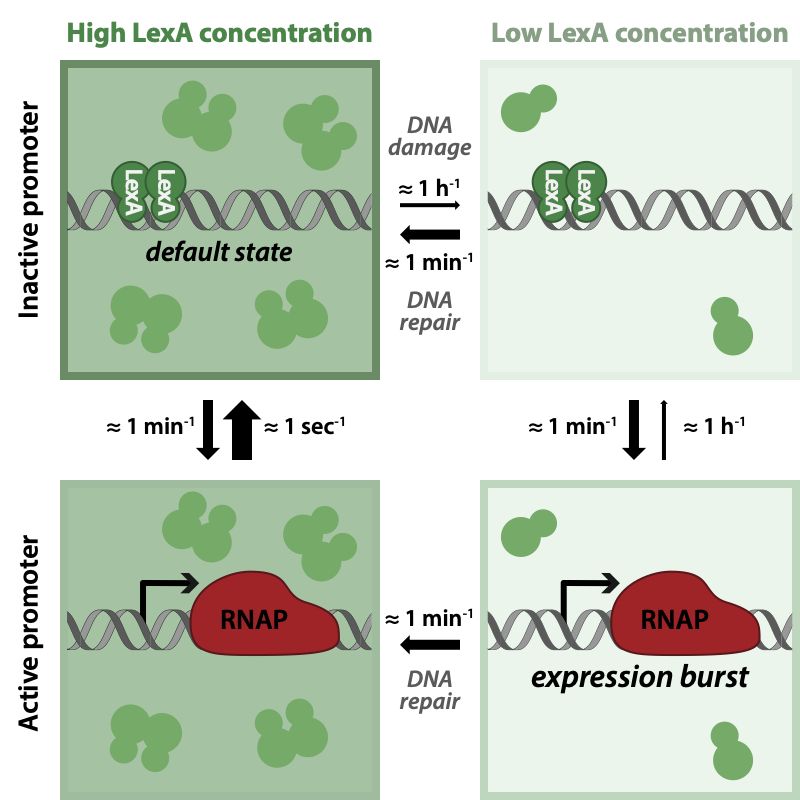
Four panels show different states of the LexA regulon with either high or low concentration of LexA and a target promoter either bound by the LexA repressor, or unbound and active. The figure also indicates the typical rates of transition between these four states. While transitions from bound to unbound always occur at a rate of about 1 per minute, rebinding is very fast when LexA is high (i.e. 1 second) and very slow when LexA is low. Therefore, expression bursts only occur when LexA levels are low. Switches to the low LexA state happen roughly once an hour when DNA damage occurs, but only last on the order of 1 minute because DNA is repaired quickly. Because this is roughly the same rate as the unbinding rate, expression bursts occur stochastically during the short time periods of low LexA levels.
Paper link and graphical abstract:
journals.aps.org/prxlife/abst...
18.07.2025 21:33 — 👍 4 🔁 0 💬 0 📌 0
Different Bacterial Genes Have Different Turn-Ons
Not all genes respond in the same way to regulation by the same molecule—a property that might enable cells to produce complex genetic responses.
Because of a lack of separation of the time scales for TF binding/unbinding and fluctuations in active TF levels, non-equilibrium gene regulation may be common in bacteria.
Check out this nice piece by @philipcball.bsky.social on our new publication in PRX Life.
physics.aps.org/articles/v18...
18.07.2025 21:30 — 👍 13 🔁 3 💬 1 📌 0
BlueSky account of the scientific group Statistical Physics of Cells and Genomes (SPCG), IFOM and U Milan. Principal Investigator (PI) Marco Cosentino Lagomarsino (MCL).
http://spcg.unimi.it/
The ECCB 25th edition will be hosted by the SIB Swiss Institute of Bioinformatics, for the 1st time in Geneva, 31.08-04.09.2026.
Discover the latest advances in genomics, structural bioinformatics, systems biology, biodiversity & beyond!
www.eccb2026.org
PhD Student in Computational Biology @ Biozentrum, University of Basel
Professor at KTH, NY Genome Center, SciLifeLab, working on functional genomics and human genetics.
Research biologist and lover of nature, mostly bacteria, insects and (lately) birds
PhD in neuroscience. A biotechnologist specialized in computer science.
I do not post at all, I am here to stay updated!
Assistant Professor @biozentrum.unibas.ch • Theoretical biophysics • Postdoc ISTAustria, PhD LMU Munich, MSc Cambridge University
www.biozentrum.unibas.ch/brueckner
Statistical Geneticist, Group leader at University of Lausanne/Unisante, father, climber, runner
PhD Students from the @biozentrum.unibas.ch 🧬🦠🧪
Bacteriophages, dormant bacteria, and bacterial immunity. Assistant Professor of Molecular Phage Biology at ETH Zürich.
Systems Pharmacology and Biology Lab at @DepBiomedicine, University of Basel, Switzerland.
We exploit metabolic weaknesses for therapeutic applications.
www.zampierilab.org
Bgee is a database at SIB and UNIL to retrieve and compare gene expression patterns in multiple animal species.
bgee.org
Theoretical Biophysicist @ TUDelft, previously Princeton, Munich, Cambridge. https://sites.google.com/view/bauergroup/home
A genomics cat, sometimes Christopher H. Browne Professor of Biology at Penn. All posts my opinion only.
The Alon lab explores design principles of biological circuits, focusing on systems medicine, aging, and healthspan.
Welcome to Maier Lab!
Our research focuses on bacterial mechanics & evolution. Here, we update you on our current research. Account run by lab members
Website: https://biophysics.uni-koeln.de
Institute for Biological Physics, University of Cologne
The SIB Swiss Institute of Bioinformatics is an internationally recognized non-profit organization dedicated to biological and biomedical #DataScience.
We provide essential resources, expertise and services and represent Swiss bioinformatics.
Studying metabolic evolution, microbial macroevolution, biospheric self-organization, and the feedbacks between Earth and life. Dad, Husband, Scientist.
https://rogierbraakman.com/
Associate Dean for data science, and Professor of Medicine and Human Genetics at the University of Chicago.