Thanks Aude!
19.07.2025 05:16 — 👍 0 🔁 0 💬 0 📌 0Nitzan Tal
@nitzantal.bsky.social
@nitzantal.bsky.social
Thanks Aude!
19.07.2025 05:16 — 👍 0 🔁 0 💬 0 📌 0Thank you Brett! 😊
14.07.2025 17:59 — 👍 0 🔁 0 💬 0 📌 0Thank you Bohdana!! 😁
14.07.2025 17:43 — 👍 0 🔁 0 💬 0 📌 0Thanks Erez! 🥰
14.07.2025 17:43 — 👍 0 🔁 0 💬 0 📌 0Thanks Romish! 🤩
14.07.2025 17:42 — 👍 0 🔁 0 💬 0 📌 0Thank you Jens! 🤗
14.07.2025 17:42 — 👍 1 🔁 0 💬 0 📌 014/
... the Kranzusch lab, AND the incredible @soreklab.bsky.social that I was so proud to call my home these last few years, it has been a blast!🥰
13/
Special thanks to:
@romihadary.bsky.social, Renee Chang, @ostermanilya.bsky.social, Roy Jacobson, @erezyirmiya.bsky.social, Nathalie Bechon,
@dinahoch.bsky.social, Miguel López Rivera, Barak Madhala, Tana Wein, @amitaig.bsky.social, ...👇

12/
I was completely in love with this project from day one, and watching it grow was such an incredible experience. 💕
📝 Read more here: tinyurl.com/j4fj9k7f
Huge thanks to everyone who made it happen - especially my brilliant collaborators 👇
11/
This study shows how structure can illuminate function in bacteria–phage warfare.🦠👩🔬🧪
And how structural convergence opens a powerful path to decode the function of mysterious ORFs in viral genomes! 🔦
10/
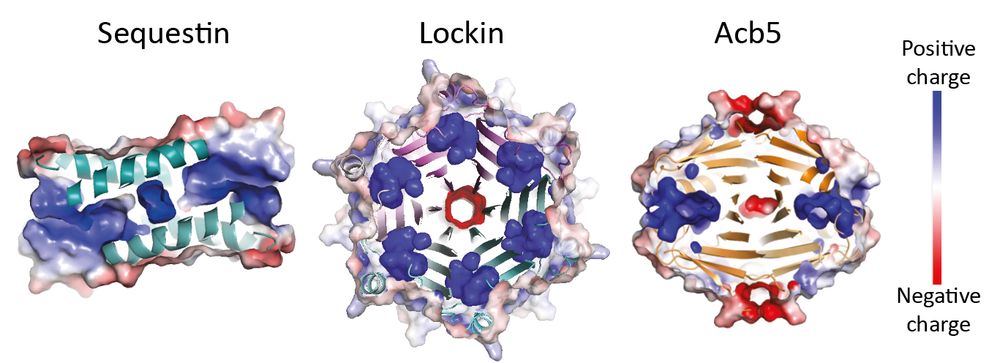
Family 3:
This one surprised us!
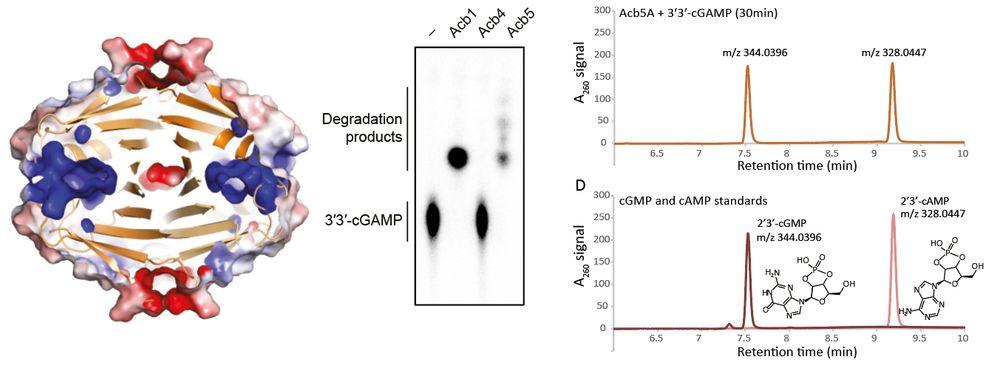
We initially thought Acb5 was a sponge, but we quickly realized it was different: it doesn’t bind the signal, it cleaves it! ✂️
This enzyme cuts cGAMP into cAMP and cGMP, depleting CBASS signaling.
A new class of viral immune evasion enzyme
9/
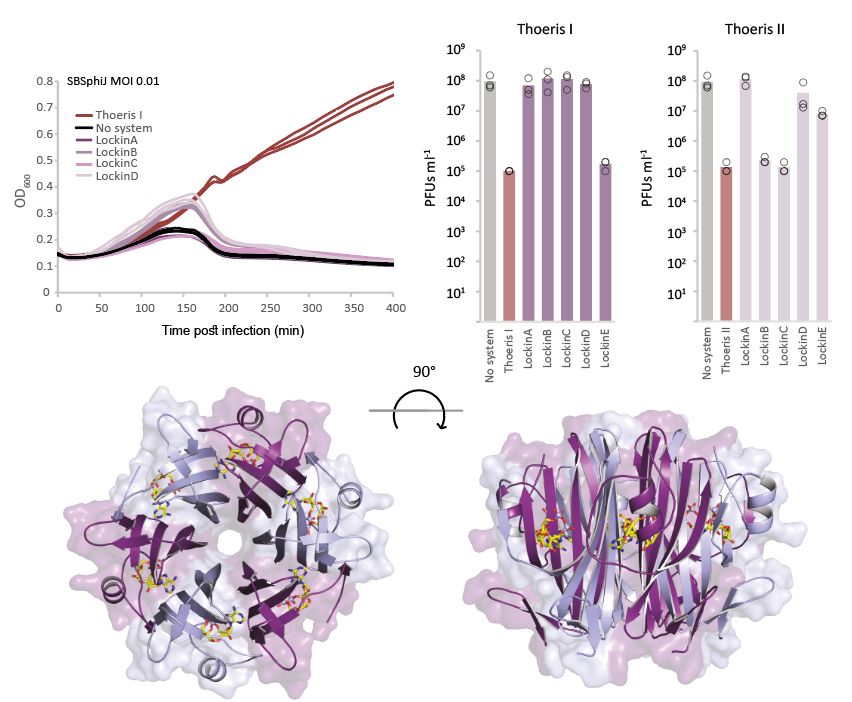
Family 2:
Lockin proteins form cog-like hexamers with deep, positively charged grooves.
We solved their structure at 1.6 Å bound to 3′cADPR, revealing not just a powerful anti-Thoeris sponge, but also an incredible structure.
It looks like a beautiful molecular flower. 🌸
8/
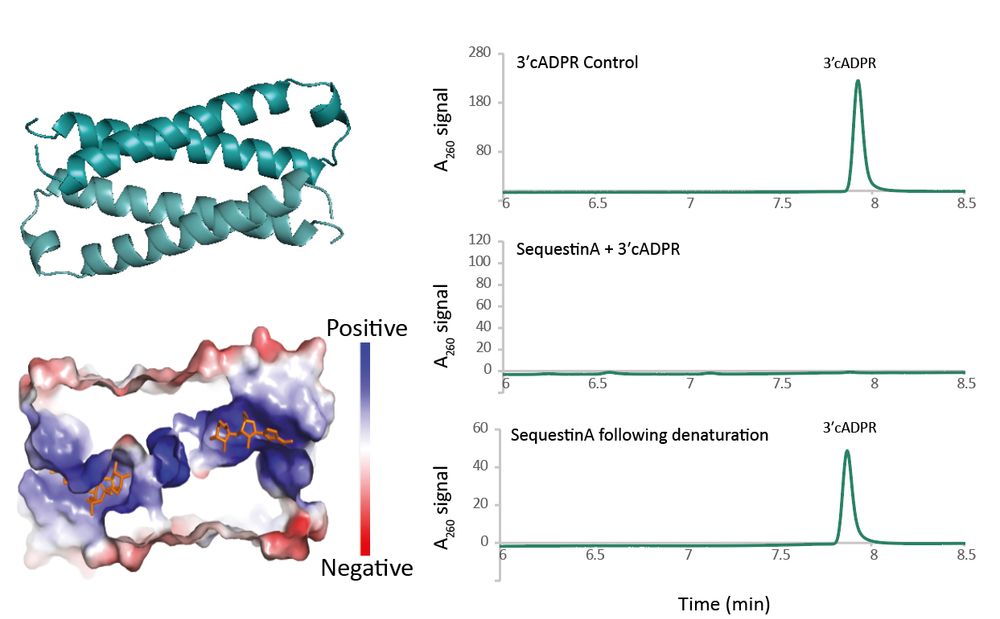
Sequestins dimerize to form binding pockets for 3′cADPR.
They’re widespread across phages, including in phage T4, where the long-uncharacterized protein Y16Q turns out to be a functional anti-Thoeris Sequestin.
7/
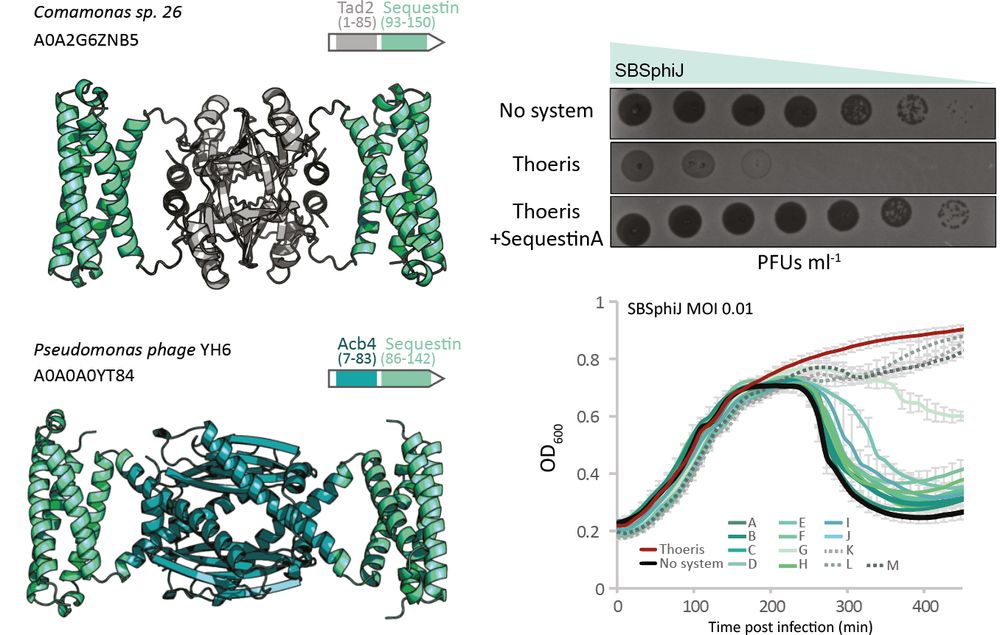
Family 1:
We first found Sequestin through a fusion-based approach: it often appeared fused to known sponges like Tad2 & Acb4 (huge shoutout to
@romihadary.bsky.social
for spotting this!)
It checked all the boxes: small, oligomeric, positively charged.
It blocks Thoeris by binding cADPR!
6/
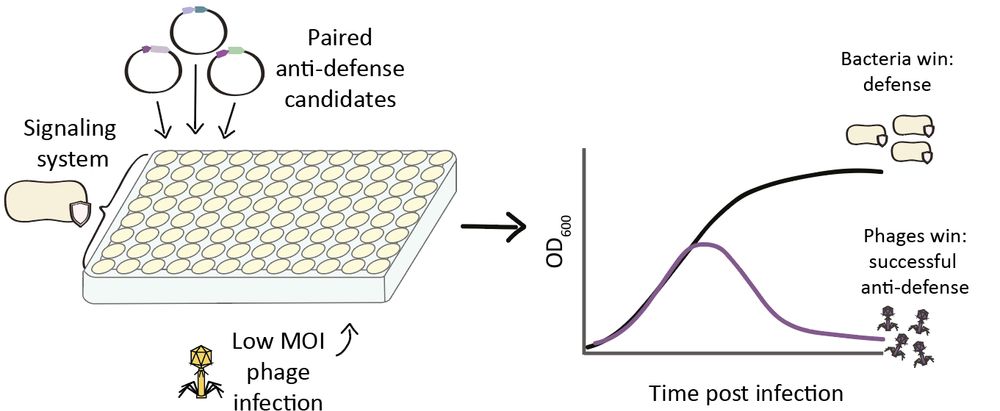
To test candidates from different families across multiple immune systems, we built a 96-well transformation and phage-infection workflow (otherwise – far too many transformations!).
This let us screen >120 proteins systematically for anti-defense activity.
5/
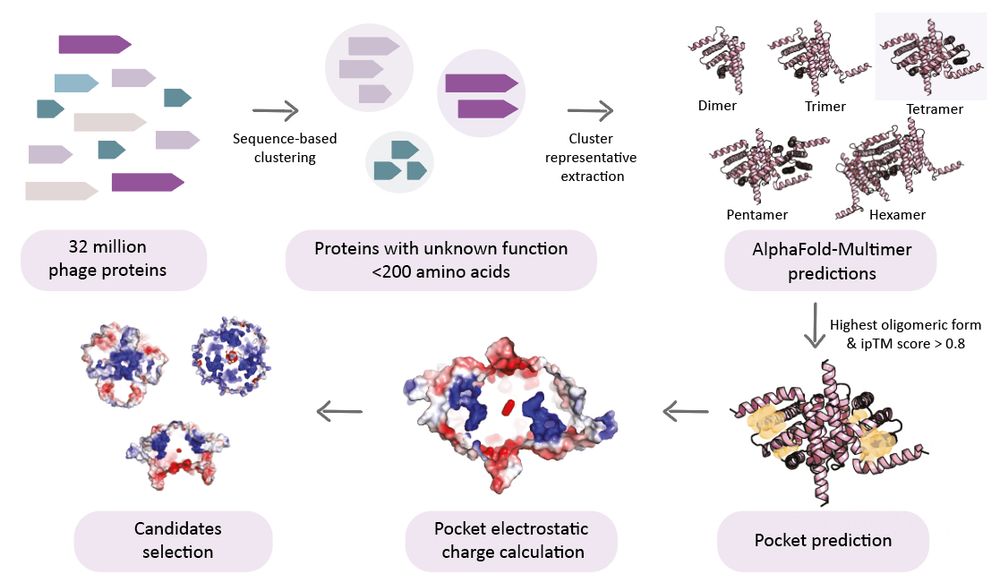
We screened ~32 million viral proteins using AlphaFold-Multimer, pocket prediction, & electrostatics filters to find candidates that look like anti-defense proteins!
We tested >120 against multiple defense systems and looked for anti-defense phenotypes.
4/
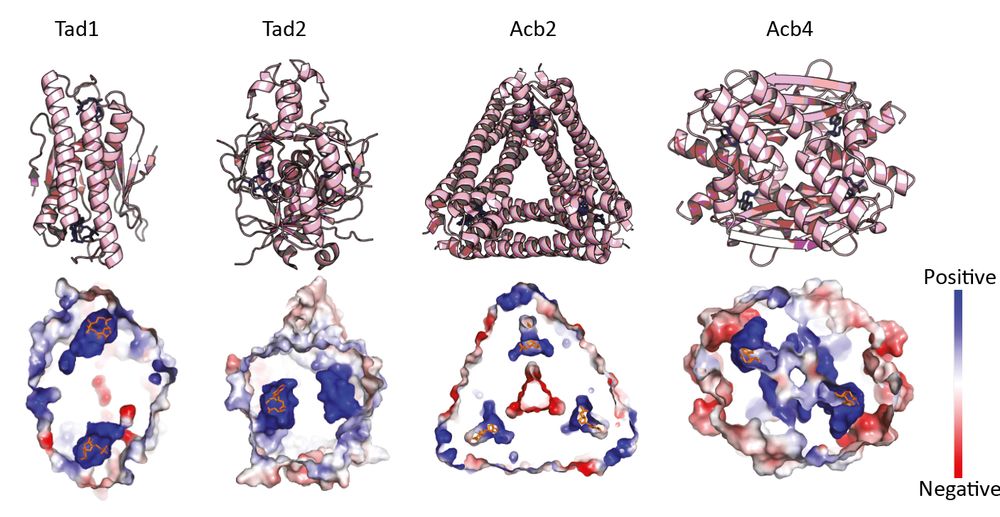
We noticed that known anti-defense sponges of signaling molecules, despite having no sequence similarity, share key structural traits:
🔹 Small size
🔹 Homo-oligomeric assembly
🔹 Positively charged pockets at protomer interfaces
We used these features to guide our search.
3/
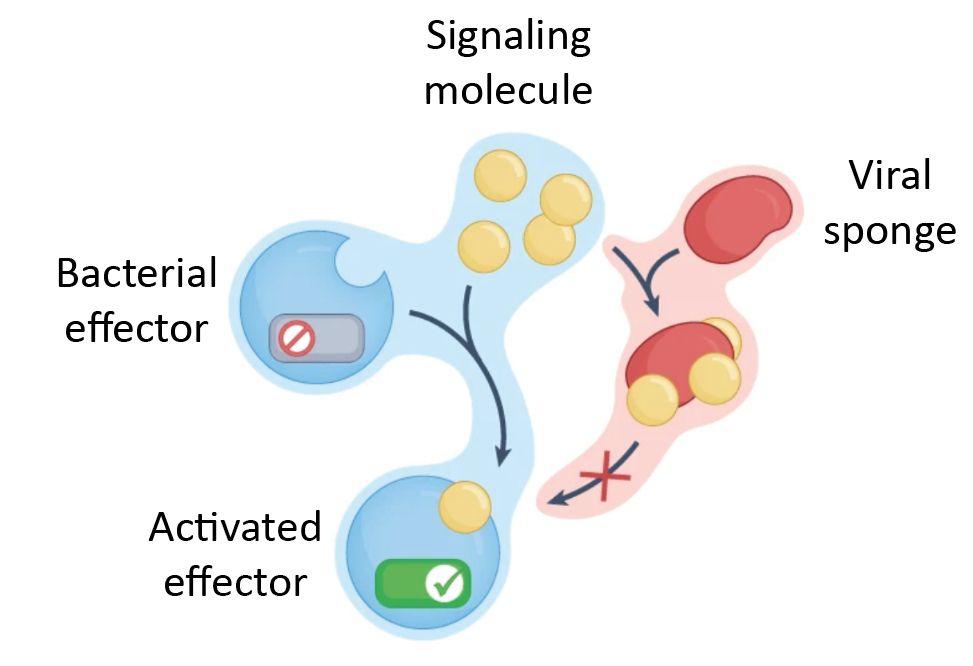
Viral sponges are especially elegant.
Instead of destroying immune signals, they soak them up - binding molecules like cGAMP or 3′cADPR and keeping the alarm from ever sounding, giving the phage a chance to win.
🖼️ Beautiful figure from Mayo-Muñoz et al.
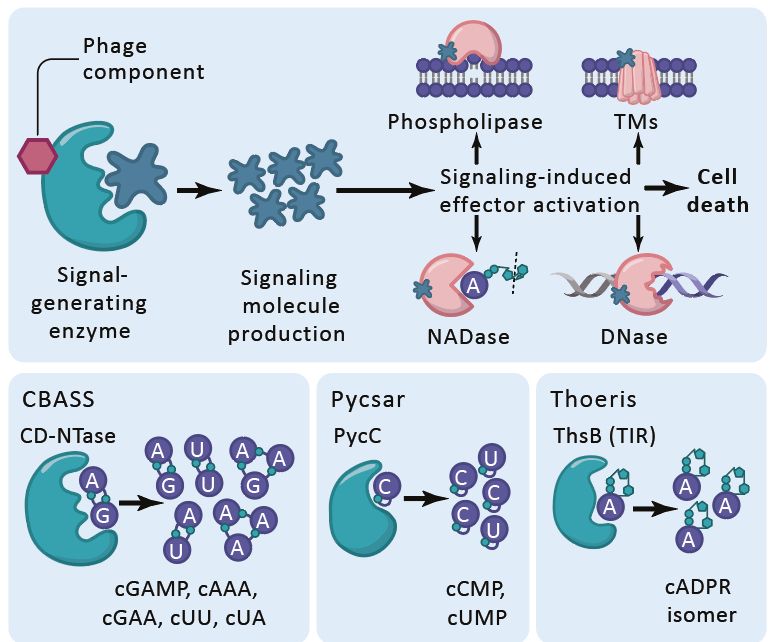
2/
Bacteria use immune systems like CBASS, Thoeris & Pycsar, which produce nucleotide messengers (cGAMP, 3′cADPR, cCMP etc.) to signal phage infection and activate immune effectors.
But phages fight back! Using proteins that bind or cleave these signals to shut down the response
📢Preprint out!
Excited to share my final work from the @soreklab.bsky.social!
We mined phage dark matter using structural features shared by anti-defense proteins (viral tools that help phages bypass bacterial immunity) to guide discovery.
Found 3 new families targeting immune signaling!