It’s out! Using cryo-ET in Dicty cells, we take a fresh in situ look at vaults. Surprisingly, we uncover vaults associated with ER and NE membranes, and find that many vaults enclose ribosomes in defined orientations, opening new avenues to their cellular function! www.biorxiv.org/lookup/conte...
16.12.2025 08:17 — 👍 106 🔁 39 💬 2 📌 5

🎉 We’re absolutely thrilled: SCALE has been selected as a DFG Excellence Cluster!
A huge thank you to our incredible team—this would not have been possible without your dedication and talent. 🙌 @dfg.de
#ClustersOfExcellence #SCALEcluster 🧵
23.05.2025 07:27 — 👍 52 🔁 19 💬 4 📌 3

(3/3) We propose a model, where the NPC dynamically acts to buffer tension applied to the nuclear envelope, thereby protecting it from potential rupture. Aberrations of the NPC scaffold would cause a reduced buffering capacity and result in NPC disintegration and potential rupture.
11.04.2025 13:26 — 👍 7 🔁 0 💬 0 📌 0

(2/3) Nup133 is a conserved component of the Y-complex, its deletion however is surprisingly is not lethal, but causes tissue and cell-type specific defects. We found that NPCs in Nup133-/- cells over-stretch and disintegrate upon induction of neuronal differentiation.
11.04.2025 13:26 — 👍 10 🔁 0 💬 1 📌 2

(1/3) Together with the labs of Valerie Doye @vdoye.bsky.social and Hans-Georg-Kräusslich, we analyzed NPC structure in Nup133 null mESCs. We found heterogenous NPCs featuring non-canonical symmetries and missing subunits. Now online @naturecellbiology.bsky.social.
www.nature.com/articles/s41...
11.04.2025 13:26 — 👍 72 🔁 22 💬 1 📌 2
Excited to share our preprint on the molecular architecture of heterochromatin in human cells 🧬🔬w/ @jpkreysing.bsky.social, @johannesbetz.bsky.social,
@marinalusic.bsky.social, Turoňová lab, @hummerlab.bsky.social @becklab.bsky.social @mpibp.bsky.social
🔗 Preprint here tinyurl.com/3a74uanv
11.04.2025 08:35 — 👍 359 🔁 141 💬 12 📌 20
New preprint on 3D heterochromatin architecture in human cells! Great collab with @sergiocruzleon.bsky.social & @johannesbetz.bsky.social from @hummerlab.bsky.social, @marinalusic.bsky.social & the Turoňová lab. Many thanks to my supervisor @becklab.bsky.social. bioRxiv: tinyurl.com/3a74uanv 🧵👇
11.04.2025 09:03 — 👍 224 🔁 67 💬 3 📌 4

Summarized, our data suggest a HIV-1 nuclear entry model where first a CypA coat around the capsid gets stripped away while the capsid is drawn into the FG-Nup mesh of NPC. Then the compression of the FG-Nups forces the NPC scaffold to crack allowing further progression of the capsid.
17.01.2025 18:43 — 👍 27 🔁 6 💬 1 📌 0
Through MD simulations we show that capsids face a significant steric barrier inside the NPC, especially when the vast amount of intrinsically disordered FG-Nups in the central channel are included. This barrier could be relieved by a crack in the NPC.
17.01.2025 18:43 — 👍 20 🔁 3 💬 1 📌 1
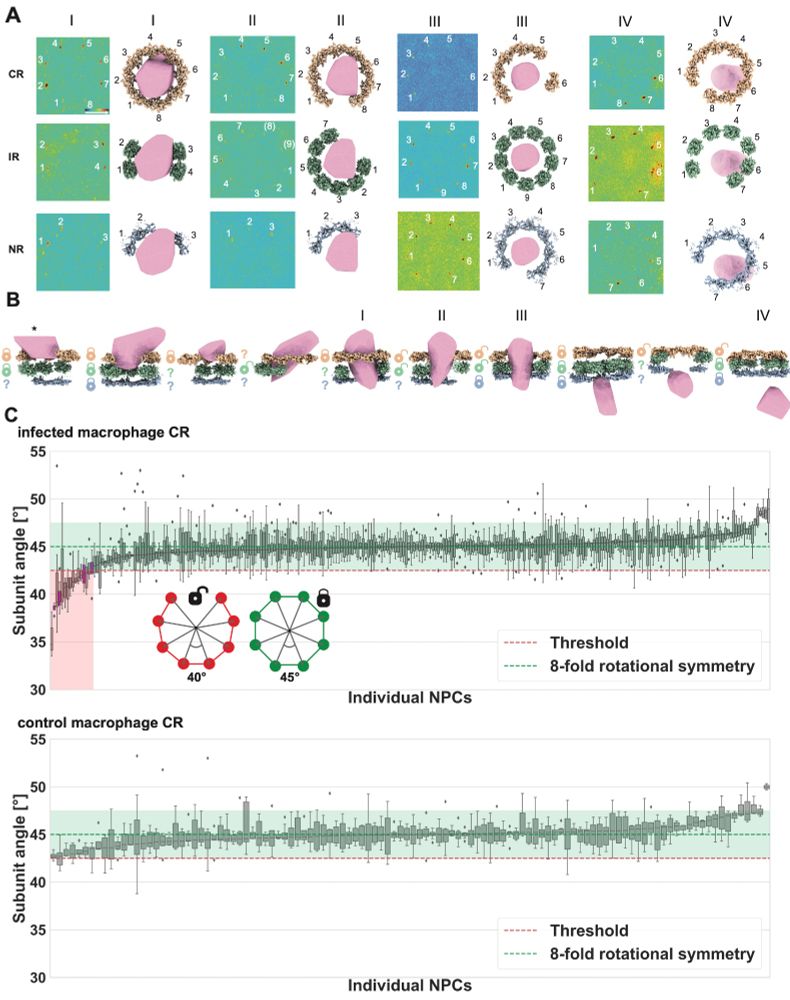
Through template matching NPC ring subunits, we were able to investigate structural changes to individual NPCs and found that capsid presence is significantly linked to NPCs being cracked open, meaning the subunit geometry exceeds the normal 8-fold rotational symmetry in the NPC scaffold.
17.01.2025 18:43 — 👍 12 🔁 0 💬 1 📌 0
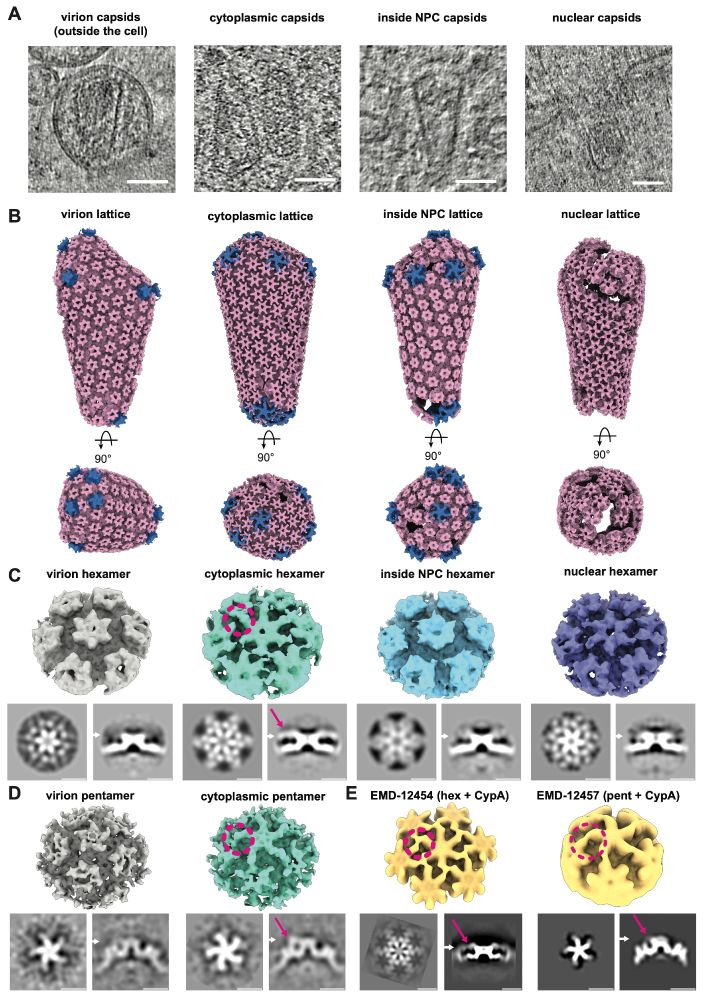
Using cryo-ET subtomogram averaging (#TeamTomo), we traced capsid (CA) lattices in virions, the cytoplasm, NPC, and nucleus. Cytoplasmic CA hexamers and pentamers showed bound density consistent with CypA, lost upon NPC entry. The CypA coat likely protects the capsid from host restriction factors.
17.01.2025 18:43 — 👍 14 🔁 0 💬 1 📌 0
CLEM-ET data showed that morphologically intact cone-shaped capsids could be found inside the central channel of NPCs and also directly underneath in the nucleus further solidifying that capsids do not need to disassemble for nuclear entry.
17.01.2025 18:43 — 👍 19 🔁 0 💬 1 📌 0

The HIV capsids were detected on the cytoplasmic side, inside the central channel, and on the nuclear side of the NPCs. These data suggest that passage through the NPC is a significant hurdle for the capsid.
17.01.2025 18:43 — 👍 13 🔁 0 💬 1 📌 0
@jpkreysing.bsky.social, @maziarheidari.bsky.social, Vojtech Zila, and others studied HIV-1 capsid nuclear entry in primary human macrophages using STED, CLEM-ET, cryo-ET (#TeamTomo), and MD simulations. First, 3D STED revealed capsid signal accumulation at nuclear pores.
17.01.2025 18:43 — 👍 16 🔁 1 💬 1 📌 1
In a great collaboration with @hummerlab.bsky.social and the Kräusslich lab: HIV capsid doesn't break at the NPC; instead, it cracks open the NPC itself! Details in Cell: authors.elsevier.com/sd/article/S... @mpibp.bsky.social @uniheidelberg.bsky.social A thread below:
17.01.2025 18:43 — 👍 429 🔁 136 💬 7 📌 22
Fresh out of the press! ✨Check out our new paper on the violent entry of the HIV capsid into the nuclear pore — and how it cracks the gate! 🤯 💥 Incredible work from @becklab.bsky.social and @hummerlab.bsky.social 🤩 More here: www.biophys.mpg.de/2842024/hivcapsid-cracks-the-npc?c=2011019 🔗 #teamTOMO
17.01.2025 16:34 — 👍 154 🔁 54 💬 1 📌 7
Hello world!
17.01.2025 17:21 — 👍 17 🔁 4 💬 1 📌 0
Evolutionary cell biology @EMBL
evonuclab.org
Full (W3) Professor Pharm. Chem. & Bioanalytics at MLU Halle-Wittenberg; Center for Structural Mass Spectrometry https://agsinz.pharmazie.uni-halle.de
https://rtg2467.uni-halle.de/
#teammassspec #massspec #ChemSky
Structural proteomics, cross-linking MS
Systems biologist • Prof at Max Perutz Labs & MedUni Vienna • dad×2 • he/him
#LoveVirology #TeamMassSpec #dataviz
📍 Vienna 🇪🇺
Biophysics of mitosis / mitotic spindle / chromosome segregation and aneuploidy
Head of Laboratory of Cell Biophysics at @institutrb.bsky.social, previously at @mpi-cbg.de, LENS Florence, Niels Bohr Institute, @hsph.harvard.edu, Uni Zagreb.
tolic.irb.hr
Research around the world of RNAs
Graduate school aiming to gain a complete molecular-level understanding of the cell! Joint program of the Max Planck Institute of Biophysics, Goethe University Frankfurt, Johannes Gutenberg University Mainz and FIAS. https://imprs-cbp.mpg.de/
PhD student @Becklab.bsky.social @Mpibp.bsky.social within @imprs-cbp.bsky.social
Professor of Molecular Biophysics&Biochemistry and of Cell Biology at Yale. Fishing all Waters.
Investigating mechanisms of eukaryotic cellular quality control with a focus on autophagy @MPIbp via #cryoET, #teamtomo, #cellbiology and #massspectrometry
Microbial cell-cell interactions | contractile injection systems | cytoskeleton | evolution | multiscale imaging | ETH Zürich
- run by lab members -
Ribosomes & Translation Regulation; Antibiotics & Resistance Mechanisms; Structural Biology & Cryo-EM; University of Hamburg.
Cell aims to publish the most exciting and provocative research in biology. Posts by Scientific Editors on the Cell editorial team. See the latest papers at https://www.cell.com/cell/
schurlab@ista.ac.at
Cryo-EM/Structural Biology/Cell Biology/Virology located @ ISTA
profile image credits: F-actin (Jesse Hansen/ISTA); photo (Anna Stöcher/ISTA)
Theoretical and computational biophysics | Postdoc at Hummer's lab
@hummerlab.bsky.social | PhD at @mpibp.bsky.social | MSc. @unistuttgart.bsky.social | Physics at @uniandes.bsky.social 🇨🇴 🇩🇪 🇪🇺
Signal transduction, systems biology and proteomics lab focusing on cancer @Lunenfeld-Tanenbaum Research Institute (LTRI), Sinai Health, Toronto, https://gingraslab.org. (Anne-Claude is also VP of Research @ Sinai Health) #TeamMassSpec
Professor in Biomolecular Mass Spectrometry and Proteomics, Utrecht University, the Netherlands














