Great response so far for the DNA Repair Meeting in April 2026 ! 🧬
A few poster spots are still available.
✨ Early-bird registration extended to February 28th
✨ New invited speakers added: check the site: dnarepairmeeting-egmond2026.com
Don’t miss this outstanding meeting!
17.01.2026 16:59 —
👍 17
🔁 8
💬 2
📌 2

Registration is open!🧬
Join us at Egmond aan Zee (April 19–24, 2026) for the next DNA Repair Meeting. We have an amazing line-up of speakers.
📅 Deadline for early registration: Jan 10, 2026
🔗 dnarepairmeeting-egmond2026.com
12.11.2025 15:04 —
👍 22
🔁 13
💬 1
📌 2
The latest work from ours and @vram142.bsky.social lab is out! True teamwork to visualize nascent chromatin with strand resolution, using a fully reconstituted system. Very proud of superstar-PhD student Bruna, and @palindromephd.bsky.social. Learning so much from Vijay’s amazing technologies! RT
21.09.2025 07:12 —
👍 36
🔁 12
💬 2
📌 0
Are you interested in working on related projects? 🌱
The Knipscheer lab at the Hubrecht Institute (Utrecht, the Netherlands) has openings for a postdoc and a PhD position.
Apply here: www.hubrecht.eu/jobs
19.09.2025 12:32 —
👍 13
🔁 10
💬 1
📌 1

Just one month left to apply for the Hubrecht International PhD Program! This fully funded 4-year PhD offers the opportunity to join a high-profile lab in molecular & developmental biology, exceptional training, and mentorship from leading scientists.
Apply before September 15 www.hubrecht.eu/hipp/
14.08.2025 10:02 —
👍 6
🔁 3
💬 0
📌 0

🧬 Looking for a PhD position in molecular or developmental biology? 🔬
✉️ The Hubrecht Institute now has its very own PhD program! Applications are open and will be accepted until September 15th. Read more here 👉 www.hubrecht.eu/hipp/
10.07.2025 11:19 —
👍 14
🔁 13
💬 0
📌 5
Thanks!
19.06.2025 11:57 —
👍 0
🔁 0
💬 0
📌 0
Thanks Sandra😍
16.06.2025 15:00 —
👍 0
🔁 0
💬 0
📌 0
Thanks Roman!
16.06.2025 15:00 —
👍 1
🔁 0
💬 0
📌 0
Thanks Gideon!
15.06.2025 12:24 —
👍 0
🔁 0
💬 0
📌 0
Thanks Floris!
13.06.2025 17:45 —
👍 0
🔁 0
💬 0
📌 0
Thanks Pablo!
13.06.2025 11:32 —
👍 1
🔁 0
💬 0
📌 0
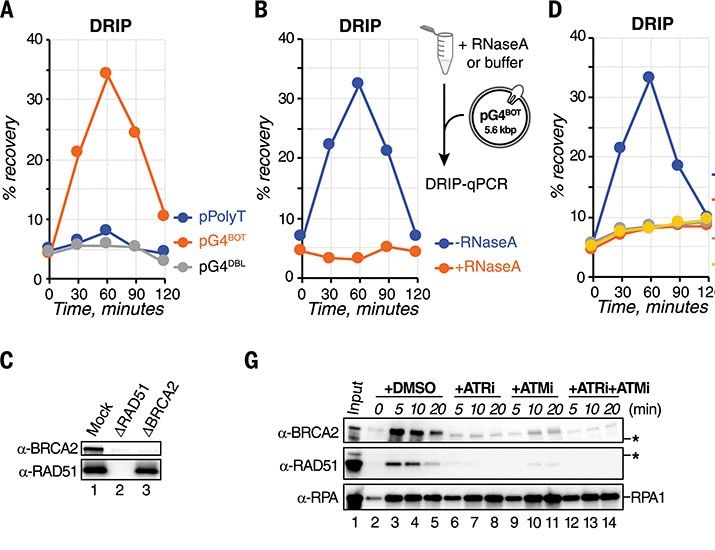
Knot good: How cells untie DNA to protect the genome
New work from the Knipscheer group reveals a mechanism involving RNA and DNA repair proteins that helps cells safely remove G-quadruplex (G4) structures.
New work from the Knipscheer group published in @science.org reveals a mechanism involving RNA and DNA repair proteins that helps cells remove G-quadruplex structures. G4 structures are knot-like formations in DNA that can cause damage if left unresolved. www.hubrecht.eu/cells-untie-dna-knots/
13.06.2025 08:37 —
👍 11
🔁 1
💬 0
📌 1
As the Science editor @dijiang319.bsky.social nicely worded: “This reveals a role for RNA transcripts in restoring genome architecture and maintaining its integrity and function”. Big thanks to all Knipscheer lab members, @hubrechtinstitute.bsky.social, and @oncodeinstitute.bsky.social (4/4)
13.06.2025 09:33 —
👍 1
🔁 0
💬 0
📌 0
Inhibition of this mechanism causes global G4 and R-loop accumulation, leading to transcription dysregulation, replication stress, and genome instability. Thanks to cells from @joost84.bsky.social from the Tijsterman lab and amazing scEdUseq by @jervdberg.bsky.social from the Oudenaarden lab (3/4)
13.06.2025 09:33 —
👍 1
🔁 1
💬 1
📌 0
We found G4s are recognized as DNA lesions leading to homology-directed invasion of RNA opposite the G4 to form a G-loop structure. This G-loop initiates G4 unwinding by DHX36 and FANCJ. Subsequent nucleolytic incision followed by renewal of the RNA-containing strand drive G-loop disassembly (2/4)
13.06.2025 09:33 —
👍 1
🔁 1
💬 1
📌 0
Knipscheer group - Hubrecht Institute
Molecular mechanism and regulation of DNA repair
Hi Erik, i am a group leader at the Hubrecht Institute and I would like to be added to the feed. Here is the link to my lab website: www.hubrecht.eu/research-gro...
13.06.2025 08:48 —
👍 1
🔁 0
💬 1
📌 0
I totally agree Helen, trying to solve a problem actually makes it worse.
12.04.2025 08:49 —
👍 1
🔁 0
💬 1
📌 0
Another day another review request (10 days). With the usual ‘suggest alternatives from underrepresented communities’. This approach doesn’t just overburden the underrepresented groups, it under-burdens the overrepresented ones, freeing them up to do stuff that is ‘objectively’ judged as ‘better’.
10.04.2025 10:55 —
👍 6
🔁 1
💬 1
📌 0
@puckknipscheer.bsky.social, @luijsterburglab.bsky.social, @marcelvanvugt.bsky.social, @sylvie-noordermeer.bsky.social, @titiasixma.bsky.social, @marcelvanvugt.bsky.social, @marteijnlab.bsky.social
09.04.2025 08:24 —
👍 0
🔁 2
💬 0
📌 0

Mark your calendar: the next Egmond DNA repair meeting will take place in 2026 from April 19 - 24!
You can pre-register to receive announcement here: forms.lumc.nl/lumc2/Egmond
Registration will open by June 2025/
23.12.2024 12:46 —
👍 21
🔁 12
💬 1
📌 0