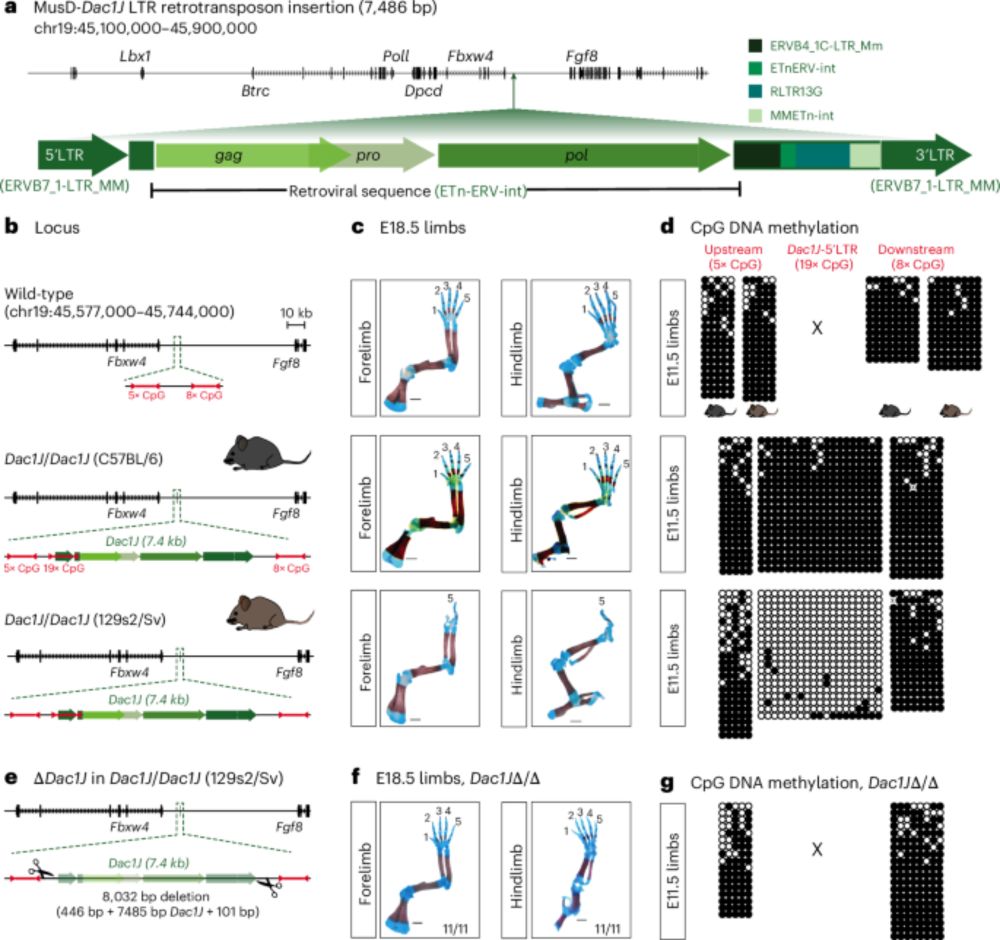
The cover of Nature Genetics July 2025 issue. Digital art of a mouse paw skeletal prep on dark background, digits I to IV disintegrating into viral particles. A caption says “An endogenous retrovirus causes limb malformation”.
Pretty surreal to see my artwork on the cover of Nature Genetics! Big congrats to @julianeg.bsky.social, @stemundi.bsky.social and the others, and thank you Juliane for letting me help bring your research to life visually.
www.nature.com/ng/volumes/5...
23.07.2025 08:06 — 👍 82 🔁 22 💬 4 📌 2
Calling all aspiring Postdocs! One extra week to apply to join our exciting HFSP-funded project to uncover how chromatin moves to function.
Deadline July 15th.
www.mdc-berlin.de/career/jobs/...
08.07.2025 09:12 — 👍 15 🔁 16 💬 0 📌 0
How do non-coding variants in enhancers lead to disease? Happy to share our recent work, led by @ewholling.bsky.social, in which we discovered that poised chromatin sensitizes enhancers to aberrant activation by non-coding mutations, contributing to disease. www.biorxiv.org/content/10.1... 1/
23.06.2025 12:56 — 👍 95 🔁 38 💬 3 📌 4

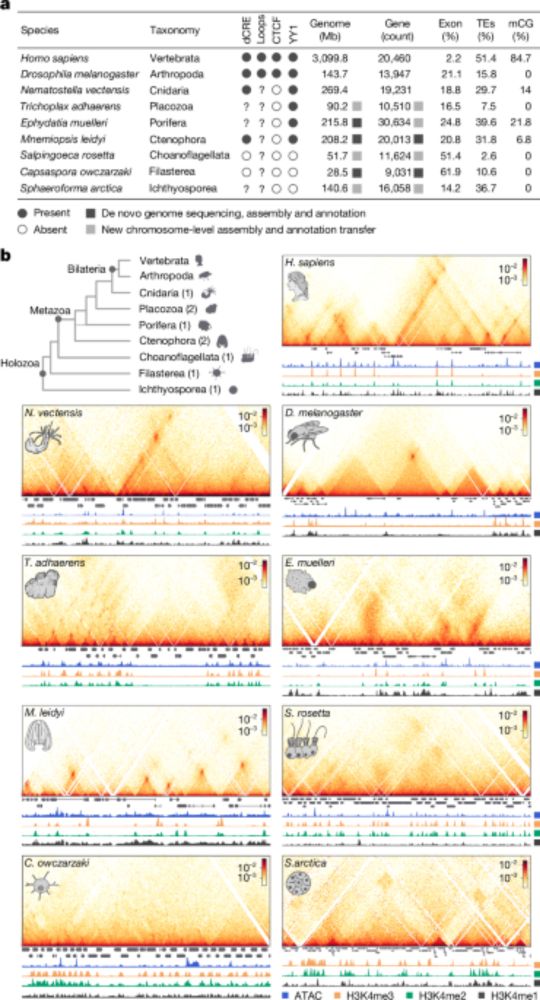
Conservation of regulatory elements with highly diverged sequences across large evolutionary distances
Nature Genetics - Combining functional genomic data from mouse and chicken with a synteny-based strategy identifies positionally conserved cis-regulatory elements in the absence of direct sequence...
How to find Evolutionary Conserved Enhancers in 2025? 🐣-🐭
Check out our paper - fresh off the press!!!
We find widespread functional conservation of enhancers in absence of sequence homology
Including: a bioinformatic tool to map sequence-diverged enhancers!
rdcu.be/enVDN
github.com/tobiaszehnde...
27.05.2025 12:19 — 👍 245 🔁 110 💬 7 📌 9
We're thrilled to continue our deep dive into how chromatin mechanics govern how our genomes function with our A-team @andersshansen.bsky.social @Davide Michieletto, and @Sandra Tenreiro.
A huge thanks to the @dfg.de and @hfspo.bsky.social for supporting us.
Stay tuned! PhDs/Postdocs coming soon!
31.05.2025 12:38 — 👍 34 🔁 5 💬 2 📌 0

portrait of a man
@drmrobson.bsky.social has won two grants from @dfg.de & @hfspo.bsky.social totaling over €2 million. He and his lab now work to decode the mechanics of #chromatin, hoping to finally define its physical state and better understand how it governs gene regulation
www.mdc-berlin.de/news/news/mi...
30.05.2025 12:50 — 👍 49 🔁 9 💬 2 📌 1
Thanks also to the @stemundi.bsky.social lab where this work started and the Robson lab where it was finished! Last, my amazing supervisor @drmrobson.bsky.social who devised the inducible DamID mouse and spearheaded the project!
08.05.2025 09:42 — 👍 2 🔁 1 💬 1 📌 0
Huge thanks to the Wunder-Kinder @imguerreiro.bsky.social, Samy & @jopkind.bsky.social without whom this project wouldn’t have happened, and to Alex and Andrea from @marionicodemi.bsky.social lab for modeling.
08.05.2025 09:42 — 👍 2 🔁 0 💬 1 📌 0

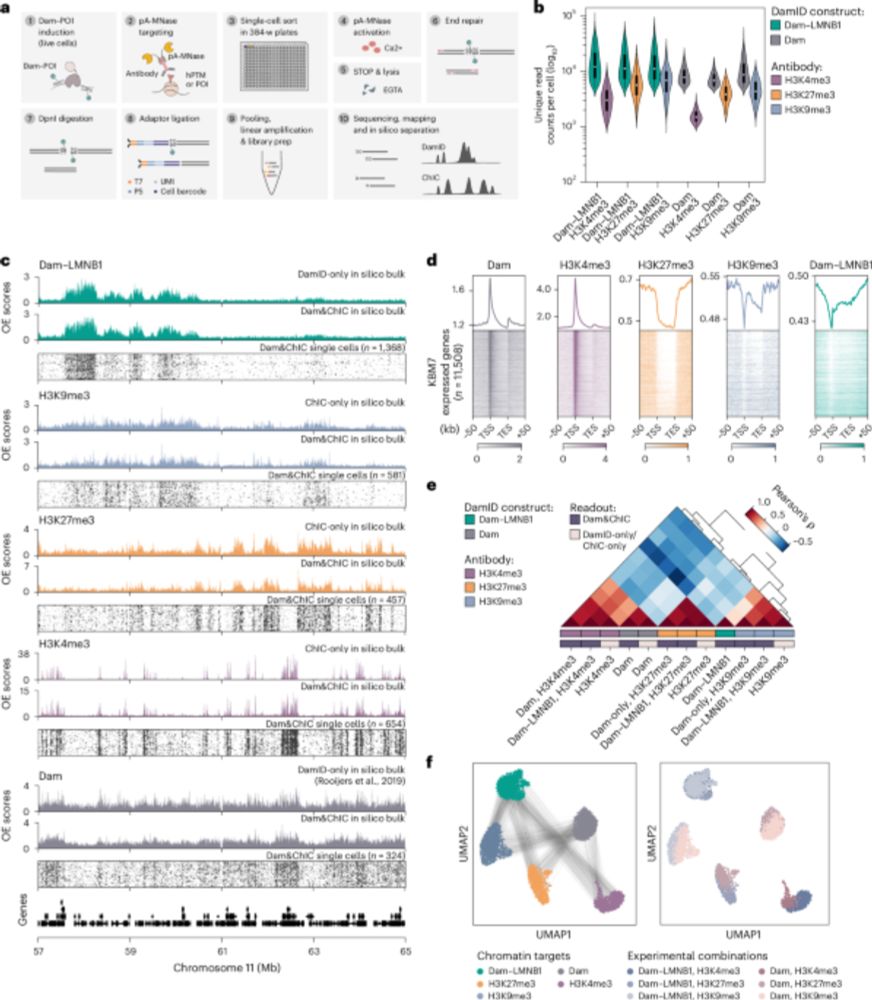
3️⃣ So what links this pre-activation repositioning to multipotency? We find it's closely tied to increased chromatin accessibility at sites bound by key limb transcription factors. This includes several factors shown to act together via a “coordinator motif” doi.org/10.1016/j.ce...
08.05.2025 09:42 — 👍 1 🔁 0 💬 1 📌 0

2️⃣ But not only genes reposition. Genes escape the periphery together with the surrounding enhancers and TADs that control their expression. Using polymer modeling & mutants, we show that CTCF boundaries and 3D chromatin topology prevent this repositioning from spreading to neighboring regions.
08.05.2025 09:42 — 👍 1 🔁 0 💬 1 📌 0
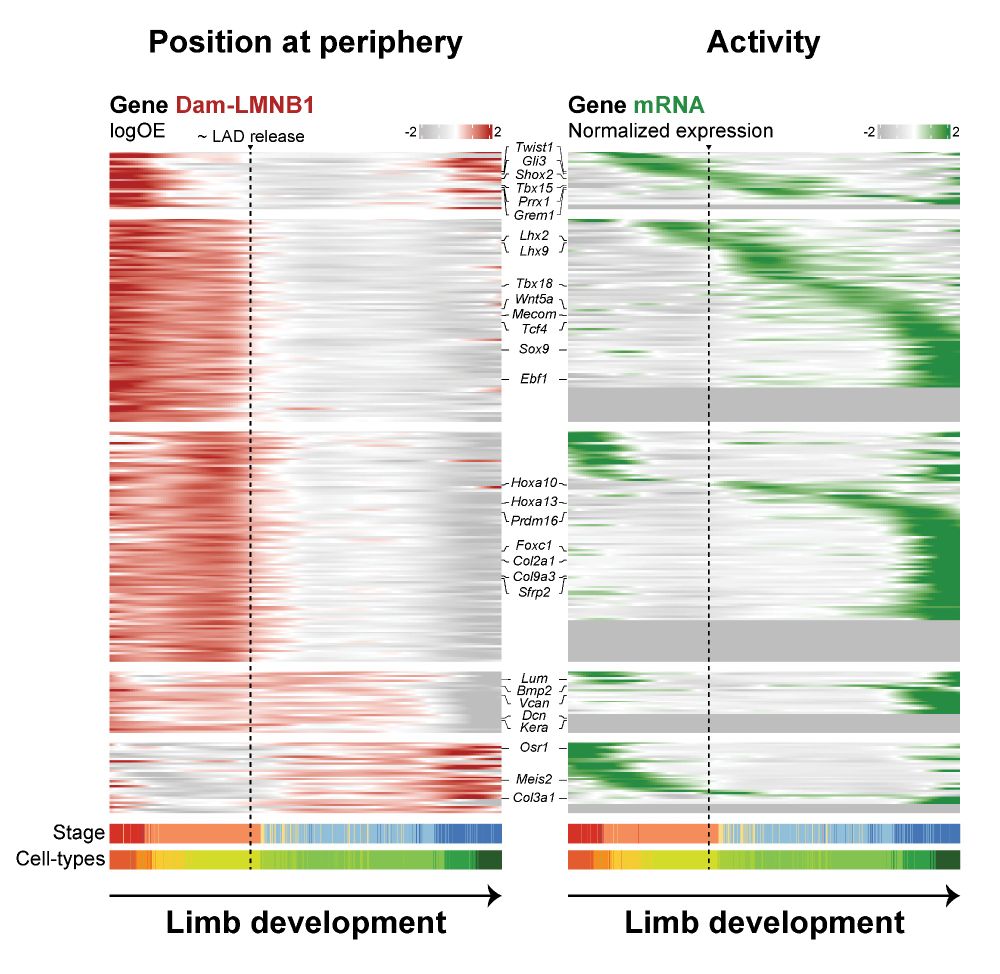
1️⃣ At the start of limb formation, hundreds of genes move away from the repressive nuclear periphery in multipotent progenitors. Amazingly, this repositioning can precede gene activation by up to four developmental days (E9.5 -> E13.5), suggesting it is a key priming step for multipotency!
08.05.2025 09:42 — 👍 1 🔁 0 💬 1 📌 0

Developmental genes have really precise expression in time and space and so must spend most of their time inactive. So how and when do they "wake up" to perform their function? Leveraging scDam&T-seq in embryonic limb development, we uncover three exciting new insights:
08.05.2025 09:42 — 👍 2 🔁 0 💬 1 📌 0

Reconfiguration of genome-lamina interactions marks the commissioning of limb cell-fates
Diverse forms of heterochromatin block inappropriate transcription and safeguard differentiation and cell identity. Yet, how and when heterochromatin is reconfigured to facilitate changes in cell-fate...
🚨 Preprint 🚨
Ever wondered how cells prepare their genomes to enable new cell-fates? In this team up with the Kind lab, we show that genes are repositioned in the nucleus to get ready for future activation and tissue formation. Read 🧵👇 to find out how and when this happens!
doi.org/10.1101/2025...
08.05.2025 09:42 — 👍 37 🔁 23 💬 1 📌 3
I am elated to share that our manuscript describing Variant-EFFECTS, a high-throughput technology we developed to precisely and quantitatively measure the effects of CRISPR-mediated edits on gene expression, is now published at @cellpress.bsky.social: authors.elsevier.com/c/1kxgiL7PXu...
17.04.2025 18:19 — 👍 96 🔁 35 💬 4 📌 4
Grateful to my wonderful collaborators worldwide:
Yuko Sato, Xingchi Yan, Simon Ulrich, @watanyatra.bsky.social, Lothar Schermelleh, Hiroshi Kimura, IrinaSolovei & my supervisor @drmrobson.bsky.social. Thank you for your hard work & support throughout! 🌍🙏
14.04.2025 20:08 — 👍 5 🔁 1 💬 1 📌 1

Is your rim staining real, or Ab-trapping? Solutions:
✅ Increase incubation time
✅ Tissue sections (antigen retrieval)
✅ Use lower-affinity antibodies/nanobodies
✅ Check cell-to-cell expression: rims only in high-expression cells = likely artifact
14.04.2025 20:08 — 👍 7 🔁 0 💬 1 📌 0

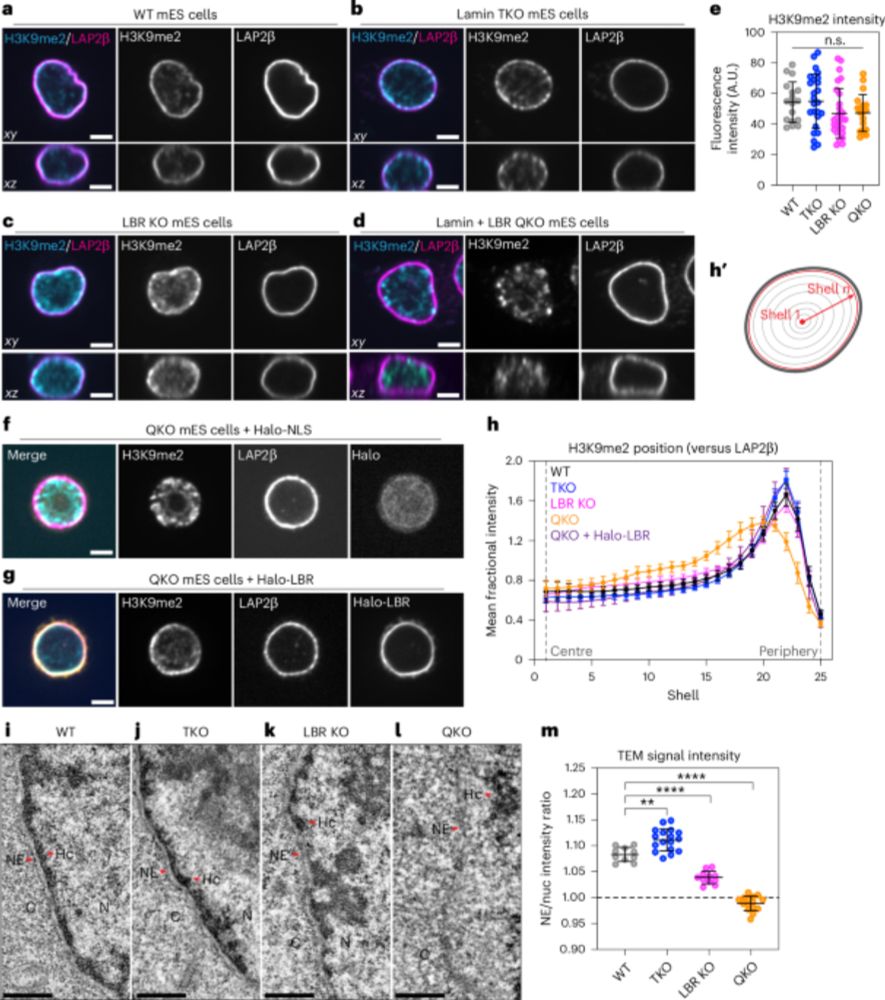
This artifact can impact any assay that relies on antibody diffusion, such as classic fluorescence microscopy, but also new-generation genomics techniques, such as CUT&Tag and CUT&RUN - SO WATCH OUT. See falsely peripheral H3K9Me2 Cut&Tag in mouse rod cells 👇
14.04.2025 20:08 — 👍 6 🔁 1 💬 1 📌 0

Ab-trapping occurs when antibodies fail to penetrate due to:
1️⃣ High epitope abundance
2️⃣ High antibody affinity
3️⃣ Low diffusion rate
Any one factor is enough! Potentially affects many antibody–epitope pairs under the right conditions. Examples 👇
14.04.2025 20:08 — 👍 4 🔁 0 💬 1 📌 0
Grateful to my wonderful collaborators worldwide:
Y. Sato, X. Yan, S. Urlich, @watanyatra.bsky.social, L. Schermelleh, @gfudenberg.bsky.social, H. Kimura, I. Solovei & my supervisor @mikerobson.bsky.social. Thanks for your hard work & support! 🤝🌍
14.04.2025 20:03 — 👍 0 🔁 0 💬 0 📌 0

Is your rim staining real, or Ab-trapping? Solutions:
✅ Increase incubation time
✅ Tissue sections (antigen retrieval)
✅ Use lower-affinity antibodies/nanobodies
✅ Check cell-to-cell expression: rims only in high-expression cells = likely artifact
14.04.2025 20:03 — 👍 0 🔁 0 💬 1 📌 0

This artifact can impact any assay that relies on antibody diffusion, such as classic fluorescence microscopy, but also new-generation genomics techniques, such as CUT&Tag and CUT&RUN - SO WATCH OUT. See falsely peripheral H3K9Me2 Cut&Tag in mouse rod cells 👇
14.04.2025 20:03 — 👍 0 🔁 0 💬 1 📌 0

Ab-trapping occurs when antibodies fail to penetrate due to:
1️⃣ High epitope abundance
2️⃣ High antibody affinity
3️⃣ Low diffusion rate
Any one factor is enough! Potentially affects many antibody–epitope pairs under the right conditions. Examples 👇
14.04.2025 20:03 — 👍 0 🔁 0 💬 1 📌 0
EPIGENETIC HULK SMASH PUNY GENOME. MAKE GENOME GO. LOCATION: NOT CENTROMERE, THAT FOR SURE
Postdoc in the Trono lab@EPFL - interested in all things noncoding and genome regulation
PhD Candidate at MDC-BIMSB with https://www.mdc-berlin.de/robson
Single-cell views of chromatin mapping🧬💻
Here for science, sparks, and fun-fact rabbit holes
Professor of Biology & Institute for Health Computing, U of Maryland; explores how viruses impact human and environmental health; 'Asymptomatic' (JHU Press, 10/2024) & 'Quantitative Biosciences' (Princeton U Press, 3/2024) & 'Science Matters' substack.
gene regulation comp bio PhD candidate @bimsb_mdc. here for finding kindred spirits & curiosities. past: applied math/cs/bio @MPI_MolGen @FU_Berlin @mit dev @soundcloud
meghan.bio
America’s Finest News Source. A @globaltetrahedron.bsky.social subsidiary.
Get the paper delivered to your door: membership.theonion.com
ICREA Research Professor. Evolution, Genetics, Neuroscience, Linguistic Cognition
Comp Bio PhD Student in Kushal Dey and Thomas Norman labs at MSK Cancer Center. Interested in gene regulation and sports
Postdoc at Stockholm University. TEs, epigenetics, bioinformatics, small RNA and Drosophila 🪰!
artemilin.dev
Postdoc @FurlongLab @embl.org - Former PhD @scicambridge.bsky.social
Interested in understanding molecular processes within the cell nucleus in the context of different cell states #chromatin #TranscriptionFactors #biophysics #technologydevelopment
PhD candidate at MDC-BIMSB, Berlin
Robson Lab - Chromatin (dys)function in disease
| BSc Biology | MSc Evolution & Behaviour |
Bioinformatics at the core — making sense of complex biological data :)
Or just “Li” |
Assist. Prof. @ Plant Bio Michigan State U. |
Also post data visualization |
Lab: https://cxli233.github.io/cxLi_lab/ |
GitHub: https://github.com/cxli233
PMGC offers high-quality, cost-effective genomics services to academic & commercial clients worldwide. Specializing in DNA seq, epigenomics, panel-based, single-cell & spatial genomics. Get in touch for custom unique solutions for your research!
PI @uam-ibmib.bsky.social, developmental epigenetics, 3D nuclear architecture, prev postdoc @bostonu.bsky.social & @ox.ac.uk 🇵🇱 🇪🇺 #StandWithUkraine 🇺🇦 https://ibmib.web.amu.edu.pl/groups/developmental-epigenetics-research-group/
Plant Science Communicator at The Sainsbury Lab, Norwich
I like all things plants 🧬🌱
Former Plantae fellow 🌟📣
Group Leader @GReD_Clermont
Chargé de Recherche @INSERM
Evolutionary biologist studying chromatin also drummer.
Jazz and epistemology. he/his
https://www.igred.fr/en/team/evolutionary-epigenomics-and-genetic-conflicts/
PhD student @Bienko Lab @Karolinska Institute and @SciLifeLab | 3D genome organization | Single-cell genomics | Method development
Biologist | UNSAM-CONICET | Bioinformatics | Cheminformatics | Genomics | Drug discovery | Diagnostics | Neglected Tropical Diseases | Argentina 🇦🇷
PhD student in the Tanenbaum lab at the Hubrecht Institute. Previously in the Pelkmans lab at UZH.
Interested in quantitative (live-cell) imaging, single-cell barcoding technologies, single-cell Omics, virus-host competition and many other things.