🫤 is the peak splitting effect reproducible with a different column?
07.07.2025 17:39 — 👍 1 🔁 0 💬 1 📌 0
Is the sample dissolved with higher organic content than your weak mobile phase?
07.07.2025 16:33 — 👍 1 🔁 0 💬 1 📌 0
With acknowledging you mean making them co-authors, right?
10.04.2025 15:10 — 👍 8 🔁 0 💬 2 📌 0
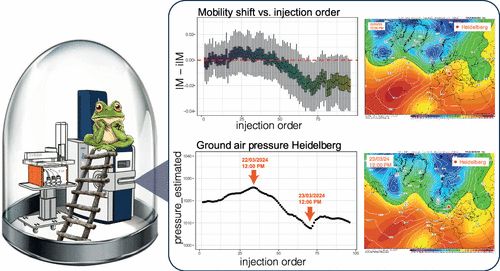
Impact of Local Air Pressure on Ion Mobilities and Data Consistency in diaPASEF-Based High Throughput Proteomics pubs.acs.org/doi/10....
---
#proteomics #prot-paper
24.01.2025 17:20 — 👍 16 🔁 3 💬 0 📌 2
I spend a bit of time talking to folk with plasma cohorts about MS vs affinity approaches for discovery. Thought it was worth a thread.
Most love the marketing. 5,000 proteins? 10,000 proteins? OLink was used in UK Biobank? Wow, mass spec is sooooo last year. Except life is not that simple.
21.11.2024 09:30 — 👍 35 🔁 16 💬 4 📌 3
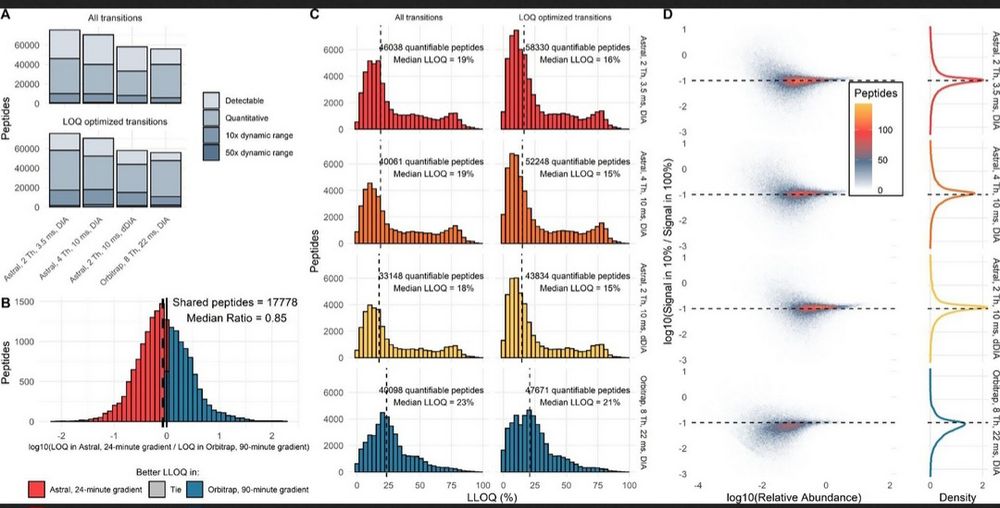
I like the matix-matched calibration curve benchmarks from one of the first papers with the Orbitrap Astral: pubs.acs.org/doi/10.1021/...
17.11.2024 16:24 — 👍 2 🔁 0 💬 0 📌 0
It also becomes tricky when studying protein synthesis regulation. Many treatments or stresses in cells cause inhibition of translation and standard normalisation approaches do not work well in such cases 😵💫
16.11.2024 07:30 — 👍 1 🔁 0 💬 0 📌 0
Proteomics software for analyzing high-resolution mass-spectrometry data.
By: https://bsky.app/profile/coxlab.bsky.social
Free to download: https://www.maxquant.org/maxquant/.
At MSAID, I am very lucky to work with incredibly intelligent people at the interface between mass spectrometry-based proteomics and artificial intelligence.
Transforming life sciences with cutting-edge #proteomics technology and solutions
Postdoc @VIBLifeSciences, @UGent, and @JNJInnovMedEMEA in the @CompOmics group. Interested in Metabolomics, Proteomics, and ML.
Director of bioinformatics at AstraZeneca. subscribe to my youtube channel @chatomics. On my way to helping 1 million people learn bioinformatics. Educator, Biotech, single cell. Also talks about leadership.
tommytang.bio.link
Our technology combine user-friendly workflows with automated processes. It empowers researchers, even without extensive expertise, to confidently use liquid chromatography mass spectrometry based proteomics in their work.
We are using mass spectrometry to study the dynamic proteome in health and disease
Postdoc in Küster lab at Technical University of Munich (TUM), Germany. Protein-nucleic acid interactions & photo-Xlinking proteomics | RNA | mass spectrometry | RNA-interacting proteomes | DNA-interacting proteomes | nucleotide-crosslinked peptides
A nonprofit research institute for advancing protein research in human health and disease | parallelsq.org
Cancer #Proteomics Researcher | Mass Spec Enthusiast | Analytical Chemist | FFPE #Phosphoproteomics | She/her | My opinions are my own
🇲🇽🇫🇷🇪🇺 Analytical chemist, specialized in #MassSpec #Proteomics. Leading the Immunopeptidomics Platform at HI-TRON Mainz / DKFZ
A mass spec proteomics user in the CAR-T therapeutic space
Account of the Saez-Rodriguez lab at EMBL-EBI and Heidelberg University. We integrate #omics data with mechanistic molecular knowledge into #opensource #ML methods
Website: https://saezlab.org/
GitHub: https://github.com/saezlab/
We are the Savitski lab located @ EMBL using and developing proteomics methods for assessing the state of the proteome
The Mann Lab is a pioneer in mass spectrometry-based proteomics. Posts represent personal views from lab members and Matthias Mann
Proteomics Researcher @MannLab - Max Planck Institute | PhD Biochemistry | Mass Spectrometry Expert | Bioinformatics | Aiming to Democratize Life Science with GenAI
Systems biologist • Prof at Max Perutz Labs & MedUni Vienna • dad×2 • he/him
#LoveVirology #TeamMassSpec #dataviz
📍 Vienna 🇪🇺
Professor in Biomolecular Mass Spectrometry and Proteomics, Utrecht University, the Netherlands
We productized proteomics services and tranformed the field. Here to discuss all things proteomics and occasionally stir the pot :)

