Research Group Leader Tenure Track - Structural Studies - LMB 2775 - Medical Research Council
Location: Cambridge. Vacancy: Research Group Leader Tenure Track - Structural Studies - LMB 2775. Closing Date: 16/03/2026, 23:55
Please spread the word: the Structural Studies Division @mrclmb.bsky.social is looking for a new tenure-track, independent group leader with an exciting plan in any area of Structural (Molecular & Cell) biology, in discovery biology and/or methods development. 🥳
mrc.tal.net/vx/mobile-0/...
16.02.2026 12:09 — 👍 79 🔁 101 💬 1 📌 1
Hey Charles! Thanks a lot!
10.12.2025 08:27 — 👍 0 🔁 0 💬 0 📌 0
Thanks Martin!!!
10.12.2025 08:26 — 👍 1 🔁 0 💬 0 📌 0
Thanks David! 💪
09.12.2025 21:46 — 👍 0 🔁 0 💬 0 📌 0
Thanks George!! 🤗
09.12.2025 21:18 — 👍 0 🔁 0 💬 0 📌 0
Star student & postdoc Kiarash Jamali is setting up his own group at the Ellison institute in Oxford. Join him for the most exciting machine learning research in structural biology and beyond!! 🤩 #ProudPI
09.12.2025 19:55 — 👍 17 🔁 7 💬 0 📌 0
Thank you! I’m excited
09.12.2025 20:09 — 👍 1 🔁 0 💬 0 📌 0
Thanks Alan!!!
09.12.2025 20:09 — 👍 0 🔁 0 💬 0 📌 0
Join ML wizard Kiarash to develop BioML methods that work
09.12.2025 17:47 — 👍 4 🔁 1 💬 0 📌 0
Kiarash is starting his research group on computational enzyme design, get in touch with him if you want to be part of it.
09.12.2025 17:40 — 👍 3 🔁 1 💬 0 📌 0
Thank you Randy! Will keep you updated :)
09.12.2025 16:27 — 👍 0 🔁 0 💬 0 📌 0
Thanks Arjen!
09.12.2025 16:27 — 👍 0 🔁 0 💬 0 📌 0
PS if the link above doesn't work, please try jamali-lab-2.pages.dev
09.12.2025 14:55 — 👍 0 🔁 0 💬 0 📌 0
Jamali Lab
Jamali Lab at the EIT Generative Biology Institute engineers new proteins with AI, biology insight, and wet-lab validation in Oxford, UK.
The GBI will be a fantastic place to do protein design research, with a large scale GPU cluster, expert wet lab biology groups, and a collaborative environment. For more information, please see jamalilab.com. If you are interested in a postdoc, please reach out, applications will be open soon.
09.12.2025 14:55 — 👍 1 🔁 0 💬 1 📌 0

GenBio DTP
Optional supporting text
I’m excited to announce that I will be starting my research group on computational enzyme design at the Generative Biology Institute, Oxford, UK (@eitoxford.bsky.social)! PhD applications are open, deadline is Jan 8th (www.chem.ox.ac.uk/genbio-dtp). Email kiarash.jamali@eit.org with any questions!
09.12.2025 14:54 — 👍 61 🔁 14 💬 9 📌 5
And just when you thought things were settling down, the enigmatic Asgard archaea have another surprise in store! Cell cycle/life cycle stage-specific internal compartments with almost no eukaryote-derived clues as to how they might function.
Beautiful tomography and microscopy - congrats all 🤩
07.11.2025 10:56 — 👍 44 🔁 11 💬 1 📌 0
Incredible work!
07.11.2025 19:42 — 👍 7 🔁 0 💬 0 📌 0

An Asgard archaeon with internal membrane compartments
Brilliant study led by @fmacleod.bsky.social and Andriko von Kügelgen. Tight collaboration with @buzzbaum.bsky.social and lab. Congrats to all authors!
www.biorxiv.org/content/10.1...
07.11.2025 10:44 — 👍 387 🔁 170 💬 10 📌 22

Wilkinson Lab
We discover and study reverse transcriptases
The Wilkinson Lab is open for science! @mskcancercenter.bsky.social
🧬We'll be finding funky new RNA biology, mainly by looking at reverse transcriptases (i.e. the Best Enzymes In The World)🧬
annnd: I'm hiring - come join! Especially postdocs and PhD students - please get in touch (NYC is great)
31.10.2025 19:00 — 👍 99 🔁 46 💬 5 📌 3

Cryo-EM maps and atomic models of the biotin-containing 3-methylcrotonyl-CoA carboxylase (MCC) complex and long-chain acyl-CoA carboxylase (LCC) complex
New lab preprint!
@zestytoast.bsky.social tagged a scarce mycobacterial protein in M. smegmatis with TwinStep but got… something? @kjamali.bsky.social's ModelAngelo built models & @martinsteinegger.bsky.social's FoldSeek IDed them as the biotin-containing MCC & LCC complexes
🧵
tinyurl.com/ukny4ptz
30.10.2025 03:21 — 👍 83 🔁 29 💬 3 📌 1
Quick Check Needed
We are looking for a computational postdoc to work with us on new optimisation algorithms to make #RELION even better. Join our bubbly team at the @mrclmb.bsky.social in Cambridge, UK. 🤗 RTs appreciated.
mrc.tal.net/vx/appcentre...
29.10.2025 09:17 — 👍 64 🔁 57 💬 2 📌 2
If you are interesting in developing new algorithms for #cryoEM as a #PhD student or as a #postdoc, then do email or DM me. We have exciting opportunities coming up.
05.10.2025 08:22 — 👍 4 🔁 5 💬 0 📌 0
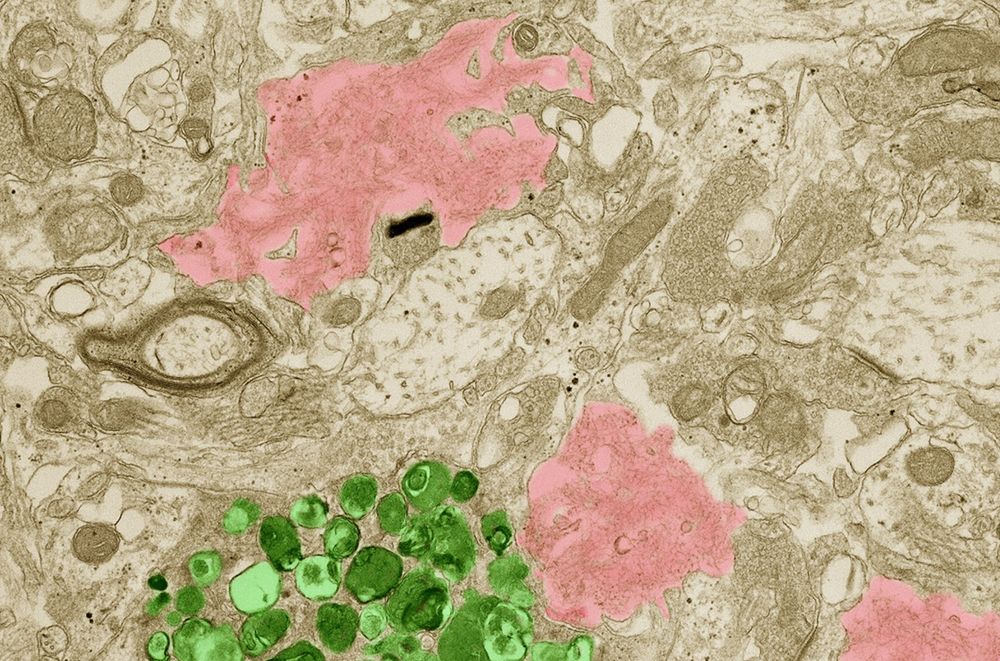
Could lithium stave off Alzheimer’s disease?
The metal is depleted in brains of people with disease and can reverse memory symptoms in mice, new study shows
Researchers found that low lithium levels are linked to Alzheimer's disease in humans, and restoring it can reverse cognitive decline in mice. For a field still searching for cure, this paper may offer a clever new approach 🧠
Read more about it in @science.org
www.science.org/content/arti...
06.08.2025 16:09 — 👍 114 🔁 34 💬 7 📌 2

Glowing E. coli bacteria in green and blues on a background of DNA bases
Rewriting the language of life: Jason Chin’s group in the LMB’s PNAC Division have synthesised E. coli with just 57 codons instead of the usual 64.
Syn57 required 101000+ codon changes & represents a bold step towards programmable life.
Read more: www2.mrc-lmb.cam.ac.uk/syn57-repres...
#LMBResearch🧪
01.08.2025 08:27 — 👍 7 🔁 4 💬 0 📌 1
Good luck Martin! I’m sure you will do great 😊
26.07.2025 01:46 — 👍 2 🔁 0 💬 0 📌 0
I am super excited to announce that I will be starting my lab at the Department of Pharmacology of the University of Zurich in Switzerland next year!
25.07.2025 07:48 — 👍 218 🔁 39 💬 23 📌 5
Very excited about our latest all-atom generative model proteina, check out the project page (research.nvidia.com/labs/genair/...) and stay tuned for the code release soon!
19.07.2025 20:46 — 👍 24 🔁 9 💬 0 📌 1
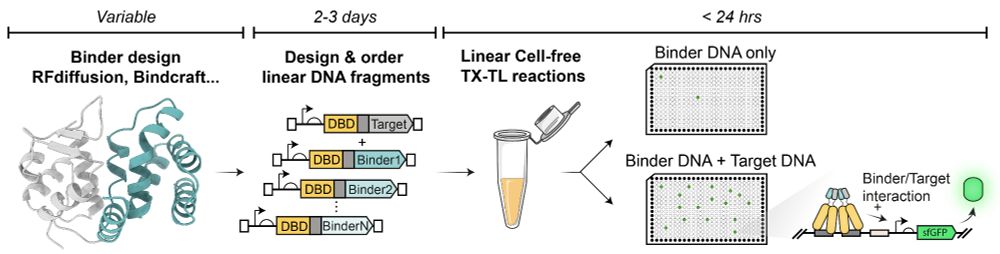
You designed binders for your favourite protein and wish there was a way to experimentally screen them within 24h w/ only a set of pipettes and a plate reader?
Check out our Cell-Free 2-Hybrid approach (CF2H)
Full post: tinyurl.com/48cz5nb6
Preprint: www.biorxiv.org/cgi/content/...
17.07.2025 11:41 — 👍 67 🔁 23 💬 0 📌 3
Professor of Microbiology @ WUR 🇳🇱 | Evolutionary microbiology | Exploring the Tree of Life | Archaea aficionado | Symbiotic interactions | husband, father, cat-person, eternal house renovator, secretly a Jedi knight
Reviewing the UK's Sticky Toffee Puddings
Automated discovery of AI x Bio papers, blogs, and news.
librarian and illustrator.
will post later, too sleepy
My shop: https://www.willquinnart.com/shop/prints
Incoming DDLS Assistant Professor, Data Science and AI
division, Chalmers University | Formerly at JHU-LangmeadLab, UNIL-DessimozLab, WUR-deRidderGroup | Pangenomics for studying genome variation and evolution | Hiring PhDs/Postdocs CGRLab.github.io
Investigating mechanisms of eukaryotic cellular quality control with a focus on autophagy @MPIbp via #cryoET, #teamtomo, #cellbiology and #massspectrometry
Global President, Ellison Institute of Technology Oxford. Senior Fellow, Worcester College, Oxford University. Former President: University of Michigan, University of British Columbia & University of Cincinnati.
Assistant Professor at the University of Virginia, Biochemist, Molecular and Structural Biologist, Husband, Father, Traveller
#cryoem #ribosome #rna
med.virginia.edu/jomaa-lab/
Assistant Professor @UT Austin | Structural Virology | cryo-EM and cryo-ET | Postdoc with John Briggs @MRC_LMB @MPI_biochem | PhD with Elizabeth Wright @GeorgiaTech @Emory
PhD student in computational Chemical Biology at Imperial College London
PhD student @Crick
Zanetti & Carlton Lab
Interested in Cell Biology, cryoET
EMDB Team Leader - EMBL-EBI. wwPDB Principal Investigator. Fathering, mixology & DIY enthusiast. #CryoEM #CryoET #firstgen. All opinions are my own.
Asst. Prof. Uni Groningen 🇳🇱
Comp & Exp Biochemist, Protein Engineer, 'Would-be designer' (F. Arnold) | SynBio | HT Screens & Selections | Nucleic Acid Enzymes | Biocatalysis | Rstats & Datavis
https://www.fuerstlab.com
https://orcid.org/0000-0001-7720-9
Investigating protein nanomachines and their dynamics.
Laboratory head at Monash University 🇦🇺.
ARC DECRA Fellow.
Structural biologist. Author of WIGGLE. He/Him. 🏳️🌈
PhD Student in the Bharat Lab at the MRC LMB
Associate Professor @ UW-Madison Biochemistry. www.limlab.org
Telomeres, Cryo-EM, Single-molecule Biophysics. Opinions are my own. 🇸🇬
Group Leader, MRC Laboratory of Molecular Biology & Joint Head of the Division of Protein & Nucleic Acid Chemistry. Senior Executive Editor, Nucleic Acids Research.
Assistant Professor, PharmTox, University of Toronto. Signalling, structural biology, protein engineering. 🇨🇦
https://caveneylab.ca
PhD student in the Jakobi lab @Bionanoscience, Delft | Imaging cool things with electrons | cryo-ET
Assist. Prof. Molecular Modelling
Ghent University
#Protein dynamics #MD simulation
Membrane proteins transporters








