If you want to implement an AI tutor, you need to study it’s performance in naturalistic settings with the kinds of context windows that student learners will provide. You can’t begin with a well formulated question about a single topic.
Students don’t know how to formulate questions well at first.
30.10.2025 10:54 — 👍 116 🔁 12 💬 4 📌 2

8 Can we live on renewables?
The red stack in figure 18.1 adds up to 195 kWh per day per person. The
green stack adds up to about 180 kWh/d/p. A close race! But please
remember: in calculating our production stack we threw all economic,
social, and environmental constraints to the wind. Also, some of our green
contributors are probably incompatible with each other: our photovoltaic
panels and hot-water panels would clash with each other on roofs; and our
solar photovoltaic farms using 5% of the country might compete with the
energy crops with which we covered 75% of the country. If we were to lose
just one of our bigger green contributors – for example, if we decided that
deep offshore wind is not an option, or that panelling 5% of the country
with photovoltaics at a cost of £200 000 per person is not on – then the
production stack would no longer match the consumption stack.
Furthermore, even if our red consumption stack were lower than our
green production stack, it would not necessarily mean our energy sums
are adding up. You can’t power a TV with cat food, nor can you feed a cat
from a wind turbine. Energy exists in different forms – chemical, electrical,
kinetic, and heat, for example. For a sustainable energy plan to add up, we
need both the forms and amounts of energy consumption and production
to match up. Converting energy from one form to another – from chemical
to electrical, as at a fossil-fuel power station, or from electrical to chemical,
as in a factory making hydrogen from water – usually involves substantial
losses of useful energy. We will come back to this important detail in
Chapter 27, which will describe some energy plans that do add up.
Here we’ll reflect on our estimates of consumption and production,
compare them with official averages and with other people’s estimates,
and discuss how much power renewables could plausibly deliver in a
country like Britain.
The questions we’ll address in this chapter are:
Is the size of the red st…
@drkatemarvel.bsky.social Are there any good current equivalents of the red stack from David MacKay 2009? It lives rent-free in my head but would like to know if it still reflects current knowledge. www.withouthotair.com/c18/page_103...
22.10.2025 19:42 — 👍 4 🔁 1 💬 1 📌 0
The science is, in fact, clear. Good studies [1] consistently show a weak association between tylenol & autism. Those studies are very careful to say they can't establish whether this is causal. A huge Swedish study used a clever sibling design to address this, and showed zero causal effect [2].
24.09.2025 18:19 — 👍 26 🔁 10 💬 2 📌 0
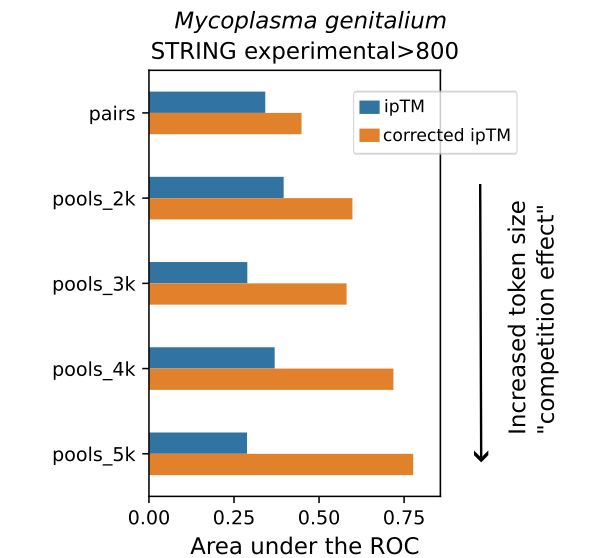
Unexpectedly, @jurgjn.bsky.social found that running Alphafold3 predictions for protein interactions can yield ipTM scores that are more predictive of true interactions when run in pools of proteins instead of pairwise predictions. Presumably, this reflects some sort of "competition effect".
22.07.2025 14:13 — 👍 87 🔁 30 💬 3 📌 3
The war on science in the US is already having an effect on private sector research like AlphaFold. Bears repeating but the private sector builds on top of things created by academic research for the public good. This hurts everyone.
28.05.2025 10:18 — 👍 239 🔁 105 💬 2 📌 12
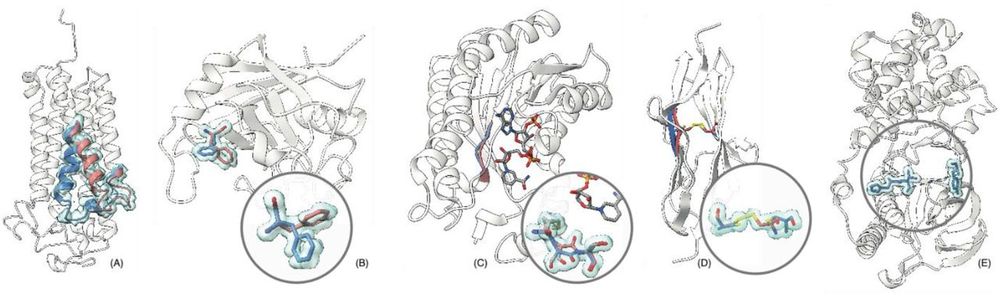
Examples of alternate conformations with environmental explanations.
(A) a protein in two states (open and closed) mediated by allosteric effects; (B) a population of two discrete rotamer states of a sidechain; (C) bound and unbound states due to ligand interaction; (D) different oxidation states of a cysteine pair. Furthermore, some regions in many protein structures exhibit such a degree of flexibility, that no accurate modelling is possible (dashed lines, E). The backbone conformation cannot be determined with crystallographic methods in such cases.
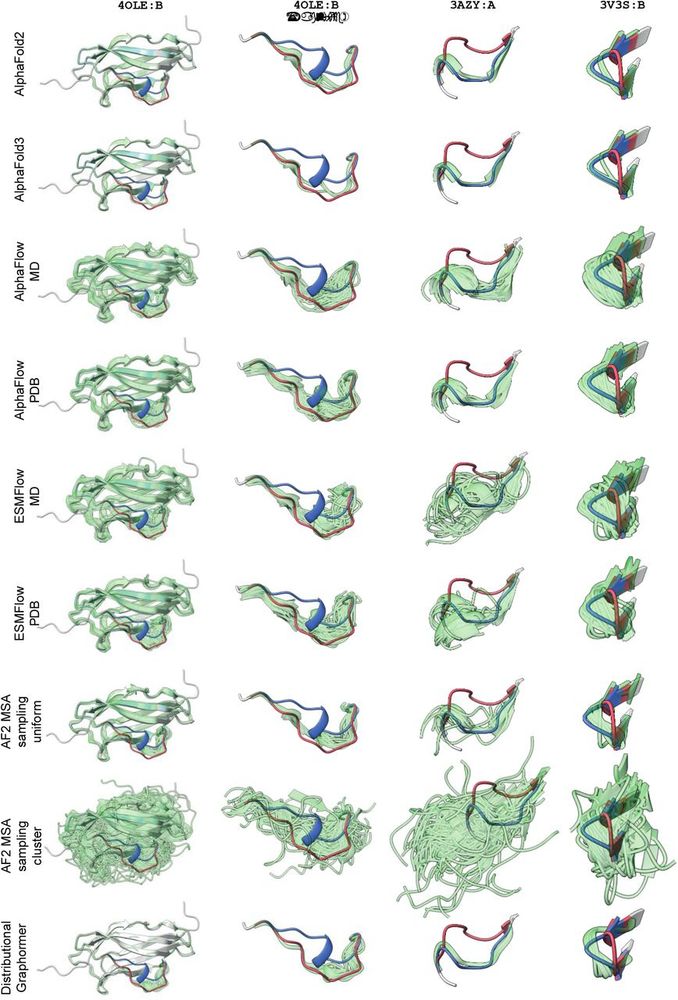
Failure of ensemble prediction algorithms in altloc segments.
Although structure and ensemble prediction techniques based on amino acid sequences are capable of accurately reconstructing the overall backbone structure (left column), they consistently fail to produce viable samples corresponding to one of the two altloc conformations (as shown in the magnified regions in the second through fourth columns).
Some crystal structures show evidence of multiple stable conformations, such as loops or side chains with two distinct regions of density. These can be correctly modeled by MD, but not modern protein ensemble prediction tools like AlphaFlow or DiG (BioEmu not tested)
www.biorxiv.org/content/10.1...
23.04.2025 14:32 — 👍 45 🔁 12 💬 0 📌 0
I miss when the biggest controversy about mRNA was whether it correlated with protein or not.
20.03.2025 13:31 — 👍 317 🔁 40 💬 10 📌 3
LLMs can't take responsibility for their mistakes. When a human journalist puts their name on AI-written text, they take on that responsibility.
Increasingly I see inaccurate and badly written news stories authored by AI, many of which have actual humans listed as authors or editors.
23.02.2025 23:56 — 👍 262 🔁 35 💬 6 📌 3
The best first sentence of a grant application I've read was (paraphrasing), "Tool X is widely used to do task Y; we will make it accessible to people living with condition Z (13% of the population) so that it is more equitable and more widely used". Let's unpack why, because there's a lesson. 🧵
05.12.2024 22:13 — 👍 223 🔁 39 💬 25 📌 5
Jurgen in our group run AF3 locally after fiddling. It worked well on a A100 and produced some bad models on a lower end GPU. We don't have access to many A100s so unless we get it working on lower-end GPUs we won't be able to use this much.
14.11.2024 19:05 — 👍 17 🔁 2 💬 2 📌 0
The former finished without errors, but the output was noise (something similar to 100% spaghetti with AlphaFold2). The latter finished with the structure posted by @pedrobeltrao.bsky.social (2/2)
15.11.2024 08:16 — 👍 1 🔁 0 💬 2 📌 0
We tried running the example from README.md (“2PV7”) on a lower-end GPU with --flash_attention_implementation=xla (described in performance.md), and on an A100 GPU without that option. (1/2)
15.11.2024 08:16 — 👍 2 🔁 0 💬 1 📌 0

BlueArk
The new blue land is waiting for you. Bring your stories with BlueArk.
1. I'm conflicted about moving old tweets to Bluesky. I like the idea in general of using @blueark.app to preserve that content.
I have a few threads posted during COVID—for example, the original analysis of the IHME model right after it came out—that seem a valuable part of the scientific record.
20.10.2024 19:48 — 👍 115 🔁 13 💬 12 📌 3
we mix informatics/biology/functional-genomics to identify new therapeutic targets/markers, for cancer and other human diseases. we ❤️ CRISPR, statistics and ML
PhD student@MIT
https://sites.google.com/view/yehlincho/home
Information about the IntFOLD server and its components. Predicted protein structures and functions from sequences. Follow for news on IntFOLD and its related methods - ModFOLD, ReFOLD, FunFOLD, MultiFOLD and ModFOLDdock.
PhD student IBG & DEU | Computational Structural Biologist
Professor detached@CNRS/Sorbonne Université, ERC and IUF fellow, co-founder@qubit_pharma, structural bioinformatics, Chemoinformatics, molecular visualization
CNRS scientist computational molecular biology. Mostly talking about life, travel, science and board games, unless it's something else.
Interested in Plant Immunity, Researcher in Kemmerling Lab, ZMBP, Tübingen, DAAD Scholar at TU Münich, MTech in IIT Kharagpur
Research Group Leader @ Izmir Biomedicine and Genome Center & DEU, TR | CASP14-15 Assembly Assessor | Computational Structural Biologist
Professor of EECS and Statistics at UC Berkeley. Mathematical and computational biologist.
Head of Estonian Biobank & Professor of Pharmacogenomics at the Estonian Genome Centre, University of Tartu.
Professor of behavioral genetics with my own twins. I mix behaviour and genetics at Tartu & McGill to understand obesity. I also mix global music as a DJ.
Scientist, Assistant Professor at MIT biology, #FirstGen
Biophysicist/Statistical physicist, Protein Quality Control / proteostasis (chaperones galore), energy-driven processes in the cell, machine learning/AI for proteins, and origins of life @EPFL, Lausanne.
Opinions are mine.
#SempreForzaToro
Research Assistant Professor, National Institute of Chemistry, Baker lab @ UW alumni, MSCA alumni. http://ljubetic-lab.si
PhD student in systems biology in the Picotti lab at ETH Zurich.
NCCR AntiResist fellow
Group Leader (RyC) at IBFG in Salamanca, Spain @CSIC.es
Fascinated about protein PTMs and how they regulate life
Cell Signalling - Chromatin - PTMs
ERC starting #PTMtalk








