Long in the making, but happy to present the Chlamydomonas chlororibosome!
Cryo-ET🔬reveals a large new domain on the small subunit, built from multiple extensions in conserved ribosomal proteins.
bioRxiv 📖: shorturl.at/q44tG
This suggests greater chlororibosome diversity than expected!
1/n 🧵
10.02.2026 08:35 — 👍 152 🔁 53 💬 4 📌 6
How does molecular valency shape condensate assembly and function? We used the CO2-fixing organelle in algae—the pyrenoid—to find out… 🧵
Preprint here!:
www.biorxiv.org/content/10.6...
@cellarchlab.com @phaips.vd.st @biologyatyork.bsky.social
#Rubisco #PhaseSeparation #Condensates #Pyrenoid
30.01.2026 15:20 — 👍 77 🔁 37 💬 1 📌 5
This one was quite the journey! The paper describing the #ChlamyDataset is finally out and on the cover of Mol Cell!
This beautiful rendering made by co-author @jessheebner.bsky.social and Holly Peterson shows an instance of mitochondrial fission found in the dataset 😍
[Maybe long thread ahead]
09.01.2026 19:14 — 👍 91 🔁 35 💬 3 📌 10
The #ChlamyDataset is on the cover of @cp-molcell.bsky.social 🖼️🥰!
Read @lifeonthewedge.bsky.social's great thread🧵 for the inside scoop🍨 on all the #TeamTomo developments already made possible since these 1829 tomos hit EMPIAR 🧪 🧶🧬
For more, here's the old preprint thread:
bsky.app/profile/cell...
09.01.2026 19:49 — 👍 36 🔁 10 💬 0 📌 0

Attention maps and PCA visualisations comparing mode
In case you're missing my poster at #NeurIPS2025 about how I fine-tuned DINOv2 to ophthalmological images, here are some animations so you don't miss out!
🔗 Preprint: doi.org/10.48550/arX...
🔗 Code: github.com/peng-lab/rmlp
🧵1/9
03.12.2025 02:18 — 👍 1 🔁 1 💬 1 📌 1
Are you also often frustrated when DINOv2 puts very high attention on background patches, rather than cute fox heads?
@virtualhomo.bsky.social found an elegant way to regularize DINOv2 training using randomised linear algebra 🤯
Check out his thread or even his poster if you're at #NeurIPS2025.
03.12.2025 08:25 — 👍 0 🔁 0 💬 0 📌 0

Randomized-MLP Regularization Improves Domain Adaptation and Interpretability in DINOv2
Vision Transformers (ViTs), such as DINOv2, achieve strong performance across domains but often repurpose low-informative patch tokens in ways that reduce the interpretability of attention and feature...
I wrote a paper with @lorenzlamm.bsky.social, @marionjasnin.bsky.social, @tingyingpeng.bsky.social, F. Eckardt and B. Schworm and got accepted at #NeurIPS2025 and fun fact: 90% of viewers are enby vegans! 🤟
So if u fit there, u might wanna check it out! Maybe also if you don't. We're allies here <3
12.11.2025 12:28 — 👍 2 🔁 2 💬 0 📌 0
I'm pleased to announce 🍦 Icecream 🍨 v0.3!
New features include:
* Training on multiple tomograms (same training time, with linear increase in RAM) 🚀
* Logging and plotting of the loss function 📉
* The --scale option is now called --eq-weight for clarity 😉
We'd love to hear your feedback! 🙏🏽
05.11.2025 14:41 — 👍 14 🔁 6 💬 1 📌 1
You might have noticed lots of activity in the napari project recently! 🚀 We're grateful for a grant from CZI that's keeping us going, but grants don't last forever: we're trying to figure out sustainable long term funding. Read our blog post to find out how you can help:
napari.org/island-dispa...
21.10.2025 14:32 — 👍 28 🔁 17 💬 1 📌 5

✨Excited that the main project of my PhD is now available as a pre-print on #bioRxiv
Here, we used #CryoET to visualise mitochondrial proteostatic stress and together with SPA #CryoEM shed light into the functional cycle of the Hsp60:10 chaperone system. #TeamTomo
🔗 www.biorxiv.org/content/10.1...
08.10.2025 15:59 — 👍 82 🔁 22 💬 2 📌 2
Time for a thread!🧵 How different is the molecular organization of thylakoids in “higher” plants🌱? To find out, we teamed up with @profmattjohnson.bsky.social to dive into spinach chloroplasts with #CryoET ❄️🔬. Curious? ..Read on!
#TeamTomo #PlantScience 🧪 🧶🧬 🌾
elifesciences.org/articles/105...
1/🧵
25.09.2025 18:00 — 👍 135 🔁 42 💬 3 📌 6
🌱 Using ‘compelling’ methods, including #CryoET, researchers mapped spinach thylakoid membranes at single-molecule precision, revealing how photosynthetic complexes are organised and settling long-standing debates on chloroplast architecture.
buff.ly/j3TSIkn
20.09.2025 13:59 — 👍 68 🔁 18 💬 3 📌 7
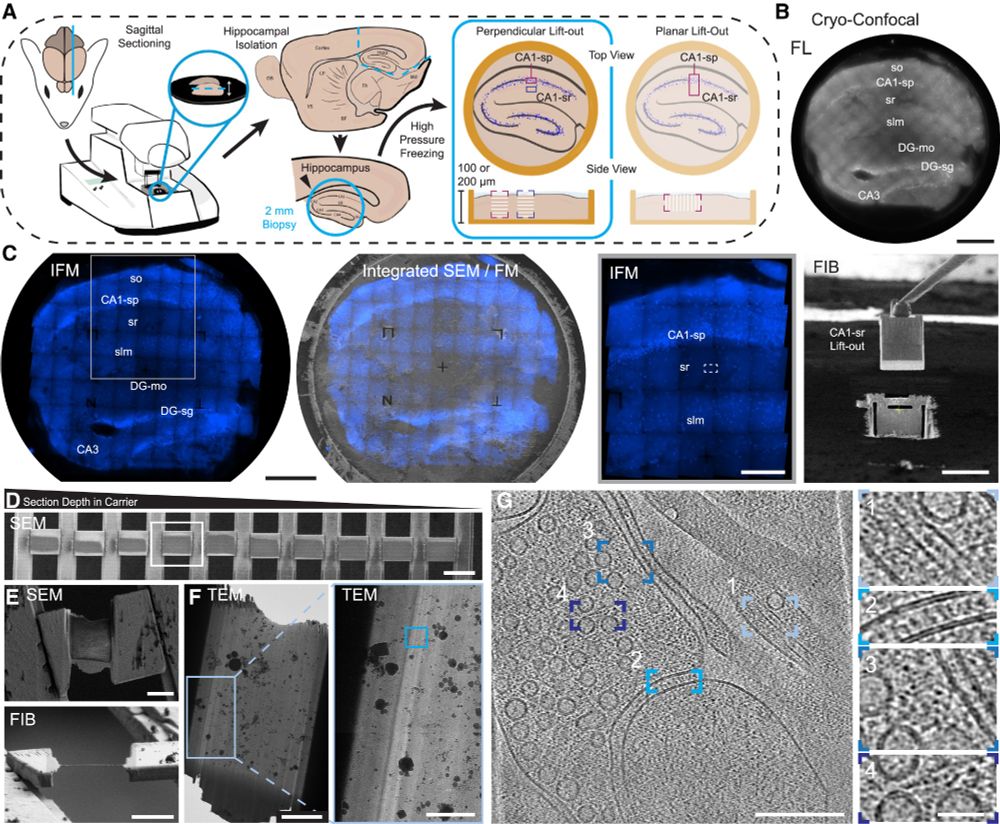
Proud to share our latest paper. doi.org/10.1016/j.cr...
Through the dedication of @glynnca.bsky.social and @cryingem.bsky.social we report a thorough method to image molecular organisation within hippocampus tissue.
Structural biology in tissue is well and truly here!
@rosfrankinst.bsky.social
17.06.2025 10:59 — 👍 62 🔁 24 💬 2 📌 4
Ooh that's awesome! Great to hear the new version improved your segmentations :)
16.06.2025 18:12 — 👍 1 🔁 0 💬 0 📌 0
Final PhD paper now reviewed and published in JSB:X. I was happy with some great reviews that improved the paper! Thanks to Sander Roet for his help with the code base, and Remco Veltkamp and @fridof.bsky.social
09.06.2025 06:56 — 👍 20 🔁 4 💬 0 📌 1

We’re kicking off the DinoSphere Online Seminar Series! Join us for our first session with Karel Mockaer (Heidelberg) & Yong Heng Phua (OIST)
📅 1 July 9AM CEST
🔗 tinyurl.com/4mjaverj
Spread the word!
@protistwtmostest.bsky.social @ehehenberger.bsky.social @chandnibhickta.bsky.social&Norico Yamada
04.06.2025 04:34 — 👍 50 🔁 30 💬 2 📌 3
You want to start tomography? Solve structures inside cells? Reach Nyquist 😳 ? @phaips.vd.st and I have a website for you! tomoguide.github.io
You'll find a tutorial on how to reconstruct tomograms, pick particles and do subtomogram averaging, using different software!
Hope it will be useful !
06.05.2025 16:42 — 👍 125 🔁 37 💬 3 📌 0
Welcome to TomoGuide
A step-by-step Cryo-ET guide
Hey #TeamTomo,
Ever been in need of a tutorial about the fundamentals of cryo-electron tomography? From preprocessing raw frames to high-res subtomogram averaging?
That's why @florentwaltz.bsky.social and I made this website!
tomoguide.github.io
Follow the thread 1 /🧵
#CryoET #CryoEM 🔬🧪
06.05.2025 16:26 — 👍 116 🔁 41 💬 5 📌 7
🚀🔬🦠 Releasing 🤖Cellpose-SAM🤖, a cellular segmentation algorithm with superhuman generalization 🦸♀️. Try it now on 🤗 huggingface.co/spaces/mouse...
paper: www.biorxiv.org/content/10.1...
w/ @computingnature.bsky.social 1/n
03.05.2025 19:12 — 👍 157 🔁 51 💬 2 📌 7

MemBrain v2: an end-to-end tool for the analysis of membranes in cryo-electron tomography
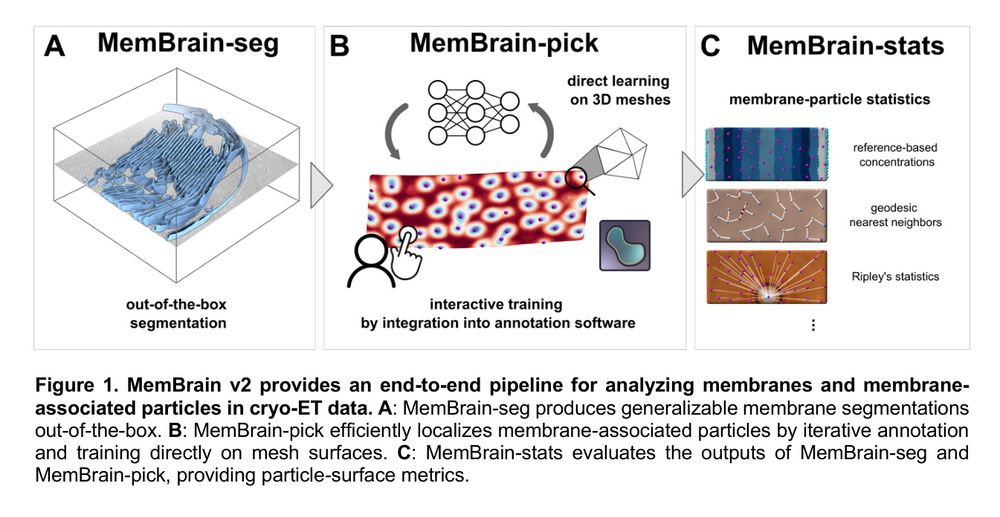
Figure 1
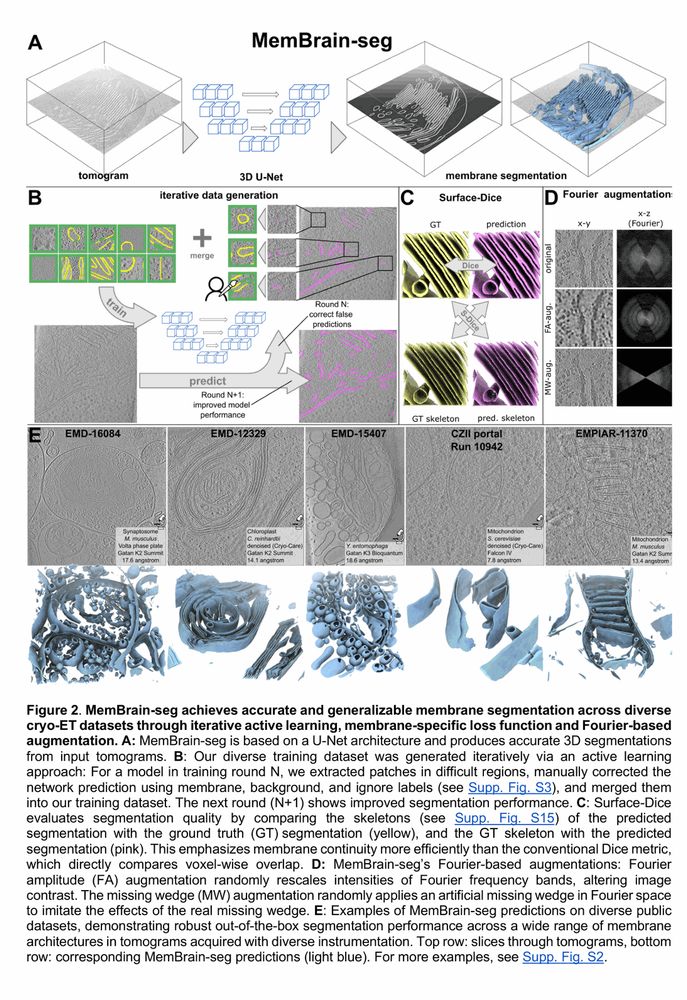
Figure 2
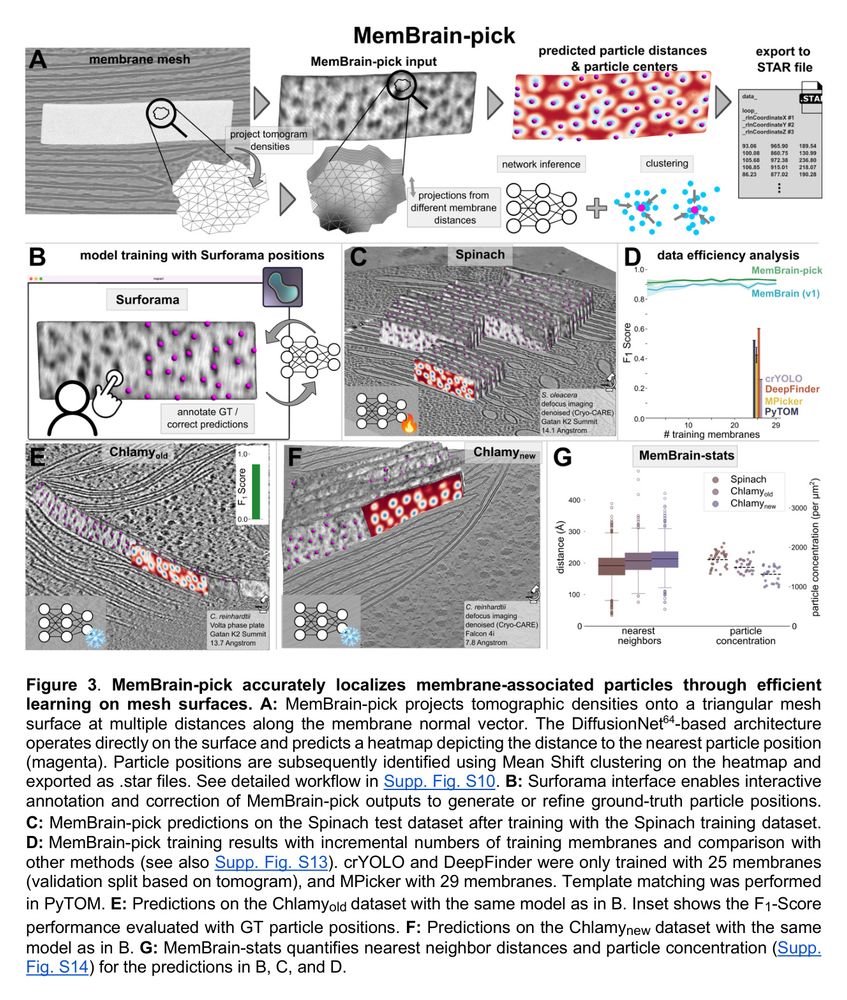
Figure 3
MemBrain v2: an end-to-end tool for the analysis of membranes in cryo-electron tomography [updated]
Cryo-ET membrane analysis via deep learning.
22.04.2025 23:21 — 👍 12 🔁 3 💬 0 📌 0
We’ve updated our powerful MemBrain-seg tool for CryoET membrane segmentation! Plus, we’re introducing two new tools: MemBrain-pick for particle picking and MemBrain-stats for statistical analysis. Feedback is warmly welcome!
25.04.2025 10:12 — 👍 15 🔁 2 💬 0 📌 0
Check out the latest version of MemBrain, spearheaded by computation superstar @lorenzlamm.bsky.social ! It can segment, pick particles and give you metrics on everything!
25.04.2025 10:25 — 👍 26 🔁 8 💬 0 📌 0
📣Huge thanks to Simon, Hanyi, @lifeonthewedge.bsky.social, @florentwaltz.bsky.social, @wojwie.bsky.social, @kevinyamauchi.bsky.social, @alisterburt.bsky.social, Ye, Antonio, Sebastian, Fabian, @ja-schnabel.bsky.social and especially @cellarchlab.com & @tingyingpeng.bsky.social for incredible support
25.04.2025 07:28 — 👍 9 🔁 0 💬 0 📌 0
🤝 Feedback
If you feel like trying one of our modules or even the full pipeline, please let us know how it goes. We are happy for any feedback and would love to improve MemBrain v2 even further to make it as helpful for the community as possible.
🧵(6/6)
25.04.2025 07:28 — 👍 6 🔁 1 💬 1 📌 0
🔑 Usability
We focused on making MemBrain v2 smooth to work with: MemBrain-seg works with a single command line, while MemBrain-pick enables data-efficient training. We facilitate the transition between modules with several Napari functionalities like the 3D lasso to crop areas of interest.
🧵(5/6)
25.04.2025 07:28 — 👍 7 🔁 1 💬 1 📌 0
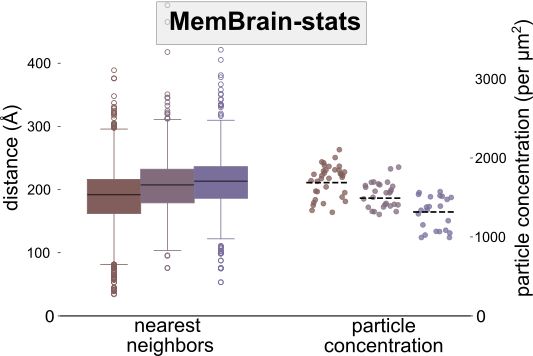
⚖️ MemBrain-stats
This module analyzes the spatial organization of particles on membranes. It takes the outputs of MemBrain-seg and MemBrain-pick to compute metrics like particle concentrations and geodesic nearest neighbor distances.
🧵(4/6)
25.04.2025 07:28 — 👍 6 🔁 1 💬 1 📌 0
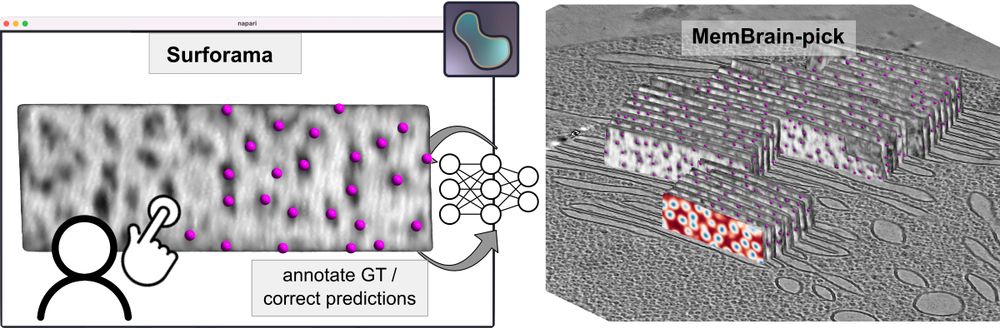
⛏️MemBrain-pick
If you’re interested in localizing membrane-associated particles, please give MemBrain-pick a try. It enables efficient training of a model to localize particles on membranes and works with the Surforama plugin for interactive annotation in Napari.
🔗 github.com/cellcanvas/s...
🧵(3/6)
25.04.2025 07:28 — 👍 5 🔁 1 💬 1 📌 0
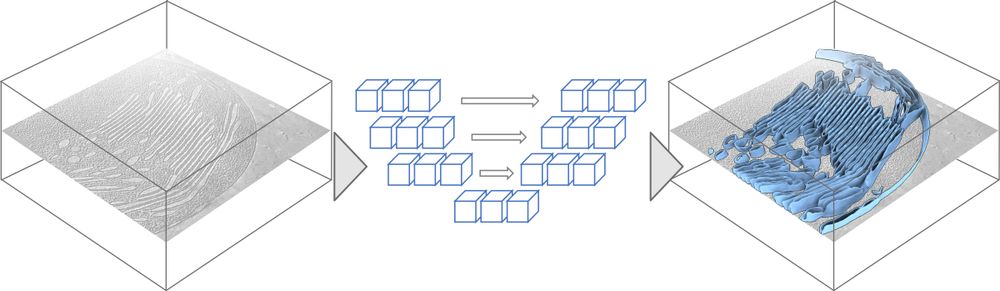
🎨 MemBrain-seg
This module allows out-of-the-box segmentation of membranes with just a single command line.
It’s based on a U-Net architecture, trained with a diverse dataset to enable generalization to many settings.
🔗 github.com/teamtomo/mem...
🧵(2/6)
25.04.2025 07:28 — 👍 5 🔁 1 💬 1 📌 0
Doctoral candidate at MDC Berlin focusing on Structural Biology | Exploring membrane protein complexes via in-situ Cryo-ET ❄️
AI/ML Research Scientist @ Bi[o]hub
Postdoctoral Researcher at BDR, RIKEN
Nonequilibrium physics of living matter Riken Hakubi research team.
Twitter: https://twitter.com/fukaity
HP: https://yfukai.net/
Human turned into a virtual homo during PhD at Helmholtz Munich
Doing my MSc in IMPRS Molecular Biology program in Göttingen
Previously at @MPI-CBG (Dr. Andre Nadler lab) and @ISTA (Dr. Alicia Michael Lab)
Assistant Professor @UT Austin | Structural Virology | cryo-EM and cryo-ET | Postdoc with John Briggs @MRC_LMB @MPI_biochem | PhD with Elizabeth Wright @GeorgiaTech @Emory
Group leader @mpi_nat, Göttingen, DE, exploring mechanisms of membrane trafficking. Cryo-EM, Cryo-ET, structural biology, cell biology.
All opinions are my own.
phd student at MDC Berlin @mdc-berlin.bsky.social
@mishakudryashev.bsky.social lab
cryo-em/teamtomo
Structural biologist, Cryo-EM aficionado, signal peptide nerd, teacher, mentor, dad, …
Machine Learning for Microscopy | Prof @ TU Dresden | https://scholar.google.com/citations?user=ZltxyqoAAAAJ
Exploring brain cell biology, organelle dynamics & proteostasis.
📍University of Cambridge @cambridge-uni.bsky.social | @UKDRI | 🔬developing live cell biosensing tech
Lab head @HHMIJanelia
neuroscience, deep learning, large-scale recordings
#Kilosort, #Suite2p, #Cellpose, #Rastermap, #Facemap
PhD candidate trying to figure out how to minimize harm and maximize the benefits of algorithms. @_kirtan_
PhD student in Structural Biology🧑🔬🌈
CryoET on mitochondria in photosynthetic organisms 🦠
Engel Lab, Biozentrum Basel
Postdoc in the Sazanov lab at ISTAustria. Using structural biology to find meaning in a world ruled by microbes. 🔬🦠
scalable data analysis, modeling, AI, software design at HHMI Janelia
https://mastodon.social/@herrsaalfeld
Science Director for AI at #RosalindFranklinInstitute creator of the #LegoBeamline and #DiamondTheGame. #SoftwareSustainabilityInstitute and #InstituteOfPhysics Fellow. #Dad he/him
🔬 studying form and function at ETH D-BSSE
🏝️ steering council and core dev of napari
https://kevinyamauchi.github.io/
We provide a cloud-native platform for cryo-EM data analysis which integrates cloud storage, scalable compute and an intuitive web-app for fast and easy end-to-end cryo-EM data analysis.



















