
Excited to share our new review/perspective on how metabolic (and cellular) context shapes enzyme evolution.
pubs.acs.org/doi/10.1021/...
@kreynoldslab.bsky.social
Scientist, teacher, mentor, mom. Definitely not the gov of IA. Interested in systems biology, molecular evolution, bacterial genomes, protein engineering, and synth bio.

Excited to share our new review/perspective on how metabolic (and cellular) context shapes enzyme evolution.
pubs.acs.org/doi/10.1021/...

After 17 years at UT Southwestern (first as a postdoc, and then starting my lab), I'm excited to be starting a new chapter in The Jenkins Department of Biophysics, at Johns Hopkins University
UTSW friends and colleagues, I will miss you very much!
New preprint! We carried out the largest complementation and inhibition study for diverse homologs in a single protein family (DHFR) to date. Highlights thread follows... 1/n 🧵
28.01.2025 01:27 — 👍 8 🔁 11 💬 1 📌 0Combining experiment and computation to tackle scientific problems in the microbial world? Consider applying for our tenure track faculty search in microbial systems biology. Application deadline coming up, Nov 29.
14.11.2024 18:08 — 👍 1 🔁 1 💬 1 📌 0
Announcing a new tenure-track faculty search in microbial systems biology! Come join a fantastic ecosystem of systems biologists and informaticians. Application deadline is Nov 29. To apply: apply.interfolio.com/154339
01.10.2024 03:17 — 👍 4 🔁 11 💬 0 📌 2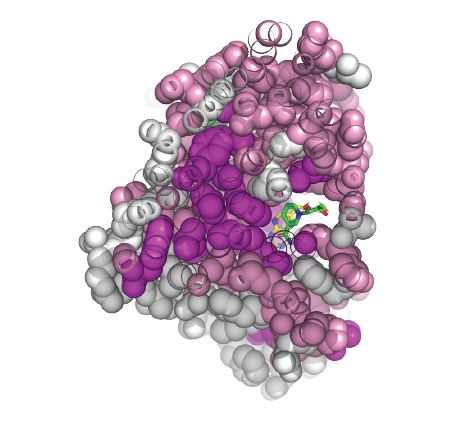
Cross section of an enzyme, color coded by mutational epistasis.
Physical protein interactions constrain interface sequence. But what about non-binding "biochemical" interactions in metabolism? We used deep mutational scanning to map sequence constraints for two enzymes that catalyze consecutive metabolic reactions: rdcu.be/dE7xl
19.04.2024 20:24 — 👍 1 🔁 0 💬 0 📌 0Our new work on (1) mapping genetic interactions in E. coli with titratable CRISPRi and then (2) using these data to build predictive gene expression -> growth rate models is out now in Cell Systems! doi.org/10.1016/j.ce...
09.02.2024 16:36 — 👍 3 🔁 0 💬 0 📌 0
Come join us! We're looking for not just one, but two postdoctoral fellows. We have one position open in biosynth pathway optimization, and another to examine how E. coli responds to perturbations in essential gene abundance. For details: www.utsouthwestern.edu/research/pos...
02.10.2023 19:47 — 👍 5 🔁 2 💬 0 📌 0
We're hiring a new faculty member in Clinical Informatics! Read more and apply at the link.
cu.taleo.net/careersectio...